Serum Paraoxonase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
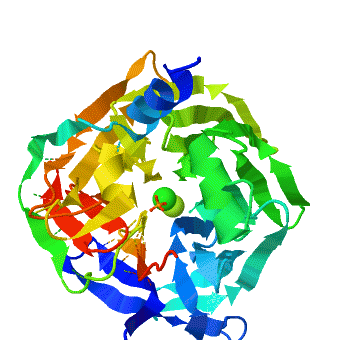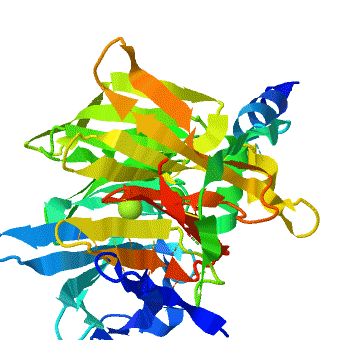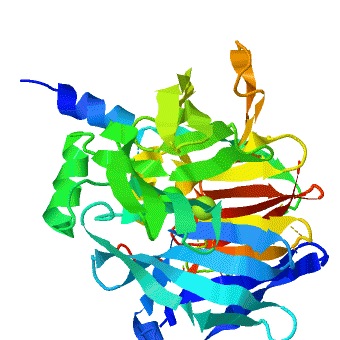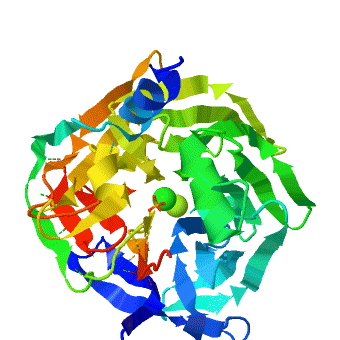
| - | <StructureSection load='1v04' size=' | + | <StructureSection load='1v04' size='450' side='right' caption='PON1 - looking down 6-bladed propellers, Ca+2 and phosphate ions seen [[1v04]]' scene='1v04/Starting_scene/3'> |
==Overview== | ==Overview== | ||
'''Serum paraoxonases''' (PONs) are a group of enzymes that play a key role in organophosphate detoxification and in prevention of atherosclerosis. There are three members in this family, PON1, PON2 and PON3, which share 60-70% nucleic acid identity. The most studied enzymes are the two isoenzymes of PON1, which differ in the residue at position 192 (Q/R). The primary activity of PON1 is lactone hydrolysis; however, this enzyme has many other activities. One of the interesting activities is the hydrolysis of organophosphates. PON1 can catalyze a variety of nerve agents such as cyclosarin, soman, etc. Therefore, it is aimed to be a nerve agent scavenger. In addition, PON1 has a role in prevention of atherosclerosis and is found to be attached to the high-density lipoprotein (HDL, “good cholesterol”). | '''Serum paraoxonases''' (PONs) are a group of enzymes that play a key role in organophosphate detoxification and in prevention of atherosclerosis. There are three members in this family, PON1, PON2 and PON3, which share 60-70% nucleic acid identity. The most studied enzymes are the two isoenzymes of PON1, which differ in the residue at position 192 (Q/R). The primary activity of PON1 is lactone hydrolysis; however, this enzyme has many other activities. One of the interesting activities is the hydrolysis of organophosphates. PON1 can catalyze a variety of nerve agents such as cyclosarin, soman, etc. Therefore, it is aimed to be a nerve agent scavenger. In addition, PON1 has a role in prevention of atherosclerosis and is found to be attached to the high-density lipoprotein (HDL, “good cholesterol”). | ||
| Line 5: | Line 5: | ||
==Structural features== | ==Structural features== | ||
| - | |||
The crystal structure of rePON1 shows<ref>PMID: 15098021 </ref><ref>PMID: 21217689 </ref> a <scene name='1v04/Six_blad/8'>six-bladed</scene> β-propeller fold. PON1 has a unique addition to the β-propeller scaffold: three <scene name='1v04/Three_helixes_pon1/9'> α-helixes</scene>, which are located on the top of the propeller. These helixes are likely to be involved in the anchoring to the [[HDL]] particles. In addition, a stabilizing <scene name='1v04/Cys_bridge/13'>disulfide bridge</scene> between Cys-42 and Cys-353 was found. The structure of rePON1 resembles that of <scene name='1v04/Loligo_v_structure/4'>''Loligo vulgaris'' DFPase</scene> (PDB [[1e1a]]). Both are six-bladed propellers with each blade consisting of four β-sheets. Moreover, in both structures two <scene name='1v04/Two_ca_ions/5'>calcium ions</scene> can be found in their central tunnel. The calcium atom, which resides at the top of the tunnel, is assigned as the ‘catalytic calcium’ (Ca-1), whereas the other calcium at the central section is assigned as the ‘structural calcium’ (Ca-2). The latter is involved in stabilization of the structure. In addition to the two calciums, there is a <scene name='1v04/Po4_ion/7'>phosphate ion</scene> , which is bound to Ca-1 in the active site. This phosphate ion is thought to come from the mother liquor. The Loligo vulgaris structure lacks the three α-helixes found in the rePON1 structure. | The crystal structure of rePON1 shows<ref>PMID: 15098021 </ref><ref>PMID: 21217689 </ref> a <scene name='1v04/Six_blad/8'>six-bladed</scene> β-propeller fold. PON1 has a unique addition to the β-propeller scaffold: three <scene name='1v04/Three_helixes_pon1/9'> α-helixes</scene>, which are located on the top of the propeller. These helixes are likely to be involved in the anchoring to the [[HDL]] particles. In addition, a stabilizing <scene name='1v04/Cys_bridge/13'>disulfide bridge</scene> between Cys-42 and Cys-353 was found. The structure of rePON1 resembles that of <scene name='1v04/Loligo_v_structure/4'>''Loligo vulgaris'' DFPase</scene> (PDB [[1e1a]]). Both are six-bladed propellers with each blade consisting of four β-sheets. Moreover, in both structures two <scene name='1v04/Two_ca_ions/5'>calcium ions</scene> can be found in their central tunnel. The calcium atom, which resides at the top of the tunnel, is assigned as the ‘catalytic calcium’ (Ca-1), whereas the other calcium at the central section is assigned as the ‘structural calcium’ (Ca-2). The latter is involved in stabilization of the structure. In addition to the two calciums, there is a <scene name='1v04/Po4_ion/7'>phosphate ion</scene> , which is bound to Ca-1 in the active site. This phosphate ion is thought to come from the mother liquor. The Loligo vulgaris structure lacks the three α-helixes found in the rePON1 structure. | ||
| Line 11: | Line 10: | ||
<scene name='1v04/Rasmol_tst_01/1'> overall structure</scene>. | <scene name='1v04/Rasmol_tst_01/1'> overall structure</scene>. | ||
| + | === Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme <ref>pmid 23318950 </ref>=== | ||
| + | |||
| + | <scene name='Journal:JMB:3/Cv/6'>Metal binding sites</scene>, especially those playing a catalytic role, exhibit high structural conservation (wildtype serum paraoxonase-1 (PON1) in the presence of either phosphate (PDB: [[3sre]]) or the lactone-analogue 2HQ (PDB: [[3srg]])). The location of the metal ion and of its ligating residues perfectly superpose, even in distant superfamily members that catalyze different chemical reactions. There exist, however, indications of changes in the configuration of catalytic metals, as part of the catalytic cycle, or upon binding different substrates. Such example is the case of the serum paraoxonase-1 (PON1), in which mutations in the H115 active site residue, induce a <scene name='Journal:JMB:3/Cv/7'>shift towards an alternative coordination mode of the catalytic Ca2+</scene> (<span style="color:royalblue;background-color:black;font-weight:bold;">wildtype PON1 is in royalblue</span>, <font color='magenta'><b>H115W mutant is in magenta</b></font> and <span style="color:salmon;background-color:black;font-weight:bold;">H115Q/H134Q mutant is in salmon</span>). PON1's native activity is the hydrolysis of lipophilic lactones, but it also promiscuously hydrolyzes organophosphates (OPs), particularly paraoxon. It uses different subsets of its catalytic machinery, and different active-site conformations, to catalyze these two reactions. However, the catalytic Ca2+, and its ligating residues are essential for both. <scene name='Journal:JMB:3/Cv/9'>H115</scene> is playing a key role in the hydrolysis of lactones. Together with E53, it activates the hydrolytic water for the lactonase activity. Further, mutations to <scene name='Journal:JMB:3/Cv/10'>Gln</scene> or <scene name='Journal:JMB:3/Cv/11'>more drastically to Trp</scene>, reduce the lactonase activity significantly (up to 600-fold). The OP hydrolase activity is, however, enhanced. To gain insight for the structural and mechanistic changes that responsible for this functional transition, the crystal structure of two H115 mutants was determined, <scene name='Journal:JMB:3/Cv/14'>H115W</scene> and <scene name='Journal:JMB:3/Cv/13'>H115Q/H134Q</scene>. These crystal structures display major rearrangements of the catalytic metal and of its ligating residues. Specifically, the <scene name='Journal:JMB:3/Cv/7'>catalytic Ca2+ has moved</scene> a 1.8 Å upwards towards the enzyme’s surface, relative to its position in the WT structure. The position of the <scene name='Journal:JMB:3/Cv/15'>structural Ca2+, however, did not change</scene>. Further, the residues coordinating the catalytic Ca2+ are also altered in the mutant structure- the side-chains of <scene name='Journal:JMB:3/Cv/16'>N168, N224 and N270 do not interact directly with the catalytic Ca2+ as in the WT structure, but through waters that are absent in the native structure</scene>. The side-chain of <scene name='Journal:JMB:3/Cv/17'>N224 also exhibits a different orientation</scene>. For <scene name='Journal:JMB:3/Cv/18'>E53, which retains its direct interaction with the catalytic Ca2+, an alternative side-chain conformation was detected</scene>. Finally, the side-chains in the vicinity of the mutations also moved, for example for the <scene name='Journal:JMB:3/Cv/19'>H115W mutant, residues H134 and L69 moved</scene> in order to accommodate the bulkiness of the Trp in position 115. The structural strudies were also complimented with biochemical, mutational and computational analysis that were in good agreement with the structural observations. The computational simulations also suggest a general base catalysis mechanism in which <scene name='Journal:JMB:3/Cv/24'>E53, possibly together with H115 and/or D269</scene>, coordinates and activates the attacking water molecule. These findings, taken together, support the notion that PON1 can accommodate <scene name='Journal:JMB:3/Cv/7'>two (or more) alternative coordination modes for its catalytic Ca2+</scene>, and that these modes may be used to catalyze different reactions. PON1's native lactonase activity occurs within the <scene name='Journal:JMB:3/Cv/21'>canonical coordination scheme</scene>, with the location of the catalytic Ca2+ being similar in PON1 and in related enzymes that are highly diverged in their sequences. The promiscuous OPH activity, however, seems to utilize a <scene name='Journal:JMB:3/Cv/23'>fundamentally different Ca2+ mode</scene>, and a different mechanism. Alongside the conformational diversity of the protein's backbone and side-chains, metal repositioning may, therefore, contribute to the catalytic versatility of enzymes and to the ease by which new enzymatic functions diverge. The shift in the Ca2+ position, from a rarely populated metal state in the WT to a dominant state in H115W, follows a general model whereby evolution capitalizes on stochastic variations, be they atomic as with PON1's alternative location of the Ca2+, or cellular (''e.g.'', transcriptional noise). Mutations do not create something from nothing. Rather, they shift the distribution such that a marginal, noise phenomenon becomes the norm. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:20, 20 November 2013
| |||||||||||
3D structures of paraoxonase
1v04 – PON + Ca + phosphate – human/rabbit/mouse/rat
2vc7 – SsPON + decanoylamino-hydroxy-dehydro-thiophenium + Co + Fe – Sulfolobus solfataricus
2vc5 – SsPON + Co + Fe
References
- ↑ Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004 May;11(5):412-9. Epub 2004 Apr 18. PMID:15098021 doi:10.1038/nsmb767
- ↑ Gupta RD, Goldsmith M, Ashani Y, Simo Y, Mullokandov G, Bar H, Ben-David M, Leader H, Margalit R, Silman I, Sussman JL, Tawfik DS. Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat Chem Biol. 2011 Feb;7(2):120-5. Epub 2011 Jan 9. PMID:21217689 doi:10.1038/nchembio.510
- ↑ Ben-David M, Wieczorek G, Elias M, Silman I, Sussman JL, Tawfik DS. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J Mol Biol. 2013 Mar 25;425(6):1028-38. doi: 10.1016/j.jmb.2013.01.009. Epub 2013, Jan 11. PMID:23318950 doi:10.1016/j.jmb.2013.01.009
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, Eran Hodis, Boris Brumshtein, Moshe Ben-David