Transfer RNA (tRNA)
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='' size='450' side='right' scene='1ehz/1ehz_default/3' caption=''> | ||
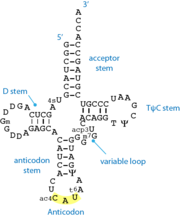
[[Image:TRNA.png|right|thumb|Standard 2D cloverleaf structure of tRNA. The shown example is methionine-specific tRNA from ''E.coli'' ]] | [[Image:TRNA.png|right|thumb|Standard 2D cloverleaf structure of tRNA. The shown example is methionine-specific tRNA from ''E.coli'' ]] | ||
'''tRNA''' or '''transfer RNA''' is stable, structured RNA present in all living cells. tRNA participates in the process of protein [[translation]] by the [[ribosome]]. Varying tRNA molecules carry a specific amino acid esterified on their 3'-OH group (the acceptor end). They also carry a specific triplet sequence, the '''anticodon''', which pairs with its complementary '''codon''' on the messenger RNA, within the ribosome. | '''tRNA''' or '''transfer RNA''' is stable, structured RNA present in all living cells. tRNA participates in the process of protein [[translation]] by the [[ribosome]]. Varying tRNA molecules carry a specific amino acid esterified on their 3'-OH group (the acceptor end). They also carry a specific triplet sequence, the '''anticodon''', which pairs with its complementary '''codon''' on the messenger RNA, within the ribosome. | ||
| Line 7: | Line 8: | ||
==Modified nucleotides== | ==Modified nucleotides== | ||
| - | + | ||
Most tRNAs contain modified nucleotides<ref>PMID:20459084</ref>, which are added post-transcriptionally by specific enzymes. Common modifications include isomerisation of uridines into pseudouridines (Ψ), methylation of either the ribose and/or the base, thiolation, reduction of uridines into dihydrouridines (D). The anticodon loop of the tRNA quite often contains hypermodified bases, the function of which is to stabilise the codon-anticodon interaction within the ribosome. The nature and position of nucleotide modifications is both specific of the organism and the tRNA type. | Most tRNAs contain modified nucleotides<ref>PMID:20459084</ref>, which are added post-transcriptionally by specific enzymes. Common modifications include isomerisation of uridines into pseudouridines (Ψ), methylation of either the ribose and/or the base, thiolation, reduction of uridines into dihydrouridines (D). The anticodon loop of the tRNA quite often contains hypermodified bases, the function of which is to stabilise the codon-anticodon interaction within the ribosome. The nature and position of nucleotide modifications is both specific of the organism and the tRNA type. | ||
Common modified nucleotides include : | Common modified nucleotides include : | ||
| Line 24: | Line 25: | ||
==Aminoacylation and function as an aminoacid carrier== | ==Aminoacylation and function as an aminoacid carrier== | ||
| - | < | + | <scene name='43/433638/Cv/2'>Glutaminyl-tRNA synthetase/tRNA complex</scene> ([[1gtr]]). |
Within the cell, each tRNA undergoes an aminoacylation-deacylation cycle. First, the cognate aminoacid is esterified on its 3'-OH by the cognate aminoacyl-tRNA synthetase. The synthetase recognizes structural features on the tRNA, which allows it to discriminate tRNA that are specific for a given aminoacid, from all other (non-cognate) tRNA. These structural features are called identity determinants. They are often (but not exclusively) located in the anticodon sequence and/or in the so-called discriminator base (position 73), just before the 3' -CCA terminus. | Within the cell, each tRNA undergoes an aminoacylation-deacylation cycle. First, the cognate aminoacid is esterified on its 3'-OH by the cognate aminoacyl-tRNA synthetase. The synthetase recognizes structural features on the tRNA, which allows it to discriminate tRNA that are specific for a given aminoacid, from all other (non-cognate) tRNA. These structural features are called identity determinants. They are often (but not exclusively) located in the anticodon sequence and/or in the so-called discriminator base (position 73), just before the 3' -CCA terminus. | ||
Once aminoacylated, tRNA associate with the elongation factor EF-Tu (bacteria) or EF1 (eucaryotes) complexed to GTP. These ternary complexes can then be recruited to the ribosome, where they go to the A-site. If a cognate codon-anticodon interaction is formed, translation can proceed, the aminoacid is incorporated within the polypetide chain and eventually, the deacylated tRNA is release for another aminoacylation-deacylation cycle. | Once aminoacylated, tRNA associate with the elongation factor EF-Tu (bacteria) or EF1 (eucaryotes) complexed to GTP. These ternary complexes can then be recruited to the ribosome, where they go to the A-site. If a cognate codon-anticodon interaction is formed, translation can proceed, the aminoacid is incorporated within the polypetide chain and eventually, the deacylated tRNA is release for another aminoacylation-deacylation cycle. | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==3D Structures of tRNA== | ==3D Structures of tRNA== | ||
===Free tRNA=== | ===Free tRNA=== | ||
| Line 80: | Line 74: | ||
* [[Ribosome]] | * [[Ribosome]] | ||
* [[2czj|tmRNA]] | * [[2czj|tmRNA]] | ||
| + | ==References== | ||
| + | <references/> | ||
| + | |||
| + | ==Reference for the structure== | ||
| + | <ref group="xtra">PMID:10943889</ref> | ||
| + | <references group="xtra"/> | ||
| + | |||
| + | |||
[[Category: Trna]] | [[Category: Trna]] | ||
[[Category: Topic Page]] | [[Category: Topic Page]] | ||
Revision as of 07:22, 30 July 2013
| |||||||||||
Contents |
3D Structures of tRNA
Free tRNA
yeast phenylalanine tRNA
human lysine tRNA (primer of HIV1 reverse transcription)
yeast aspartic acid tRNA
E. coli initiatior methionine tRNA
tRNA fragments
Complexes with aminoacyl-tRNA synthetases
Complexes with elongation factors
Complex with RNAse P
Complexes with the ribosome
See Also
References
- ↑ Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010 Jun 22;49(24):4934-44. PMID:20459084 doi:10.1021/bi100408z
Reference for the structure
- Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000 Aug;6(8):1091-105. PMID:10943889
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Wayne Decatur, Michal Harel, Frédéric Dardel, Ann Taylor, Joel L. Sussman, Alexander Berchansky
Categories: Trna | Topic Page | Translation | Modification | RNA | Amino acid