MDM2
From Proteopedia
| Line 1: | Line 1: | ||
| - | {{STRUCTURE_1ycq| PDB=1ycq | SIZE=400| SCENE= |right|CAPTION=Frog MDM2 complex with p53 transactivation domain, [[1ycq]] }}[[Image:MDM2 Header.png|thumb|left|250px|Structure of MDM2, [[1ycq]]]]The '''MDM2''' oncoprotein is a cellular inhibitor of the [[P53|p53 tumor suppressor]] in that it can bind the transactivation domain of p53 and downregulate its ability to activate transcription. In certain [[Cancer|cancers]], MDM2 amplification is a common event and contributes to the inactivation of p53. The crystal structure of the 109-residue amino-terminal domain of MDM2 bound to 109-residue transactivation domain peptide of p53 revealed that MDM2 has a <scene name='Matt_Routh_sandbox/Hydrophobic_cavity/1'>deep hydrophobic cleft</scene> on which the p53 peptide binds as an amphipathic alpha helix. The interface relies on the steric complementarity between the MDM2 cleft and the hydrophobic face of the p53 alpha helix and, in particular, on a triad of p53 amino acids-Phe19, Trp23, and Leu26-which insert <scene name='Matt_Routh_sandbox/Connection_to_p53/5'>deep inside</scene> the MDM2 cleft. These same p53 residues are also involved in transactivation, supporting the hypothesis that MDM2 inactivates p53 by concealing its transactivation domain. The structure also suggests that the amphipathic alpha helix may be a common structural motif in the binding of a diverse family of transactivation factors to the TATA-binding protein-associated factors. | + | {{STRUCTURE_1ycq| PDB=1ycq | SIZE=400| SCENE= |right|CAPTION=Frog MDM2 complex with p53 transactivation domain, [[1ycq]] }}[[Image:MDM2 Header.png|thumb|left|250px|Structure of MDM2, [[1ycq]]]]The '''MDM2''' oncoprotein is a cellular inhibitor of the [[P53|p53 tumor suppressor]] in that it can bind the transactivation domain of p53 and downregulate its ability to activate transcription. In certain [[Cancer|cancers]], MDM2 amplification is a common event and contributes to the inactivation of p53. The crystal structure of the 109-residue amino-terminal domain of MDM2 bound to 109-residue transactivation domain peptide of p53 revealed that MDM2 has a <scene name='Matt_Routh_sandbox/Hydrophobic_cavity/1'>deep hydrophobic cleft</scene> on which the p53 peptide binds as an amphipathic alpha helix. The interface relies on the steric complementarity between the MDM2 cleft and the hydrophobic face of the p53 alpha helix and, in particular, on a triad of p53 amino acids-Phe19, Trp23, and Leu26-which insert <scene name='Matt_Routh_sandbox/Connection_to_p53/5'>deep inside</scene> the MDM2 cleft. These same p53 residues are also involved in transactivation, supporting the hypothesis that MDM2 inactivates p53 by concealing its transactivation domain. The structure also suggests that the amphipathic alpha helix may be a common structural motif in the binding of a diverse family of transactivation factors to the TATA-binding protein-associated factors.<ref name="Kussie">PMID:8875929</ref> |
| - | <ref name=" | + | |
==Role in Cancer== | ==Role in Cancer== | ||
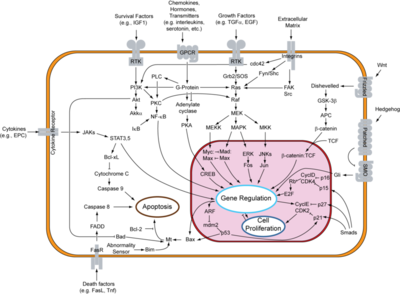
[[Image:800px-Signal transduction v1.png|Apoptosis signal pathway|400 px|thumb]] | [[Image:800px-Signal transduction v1.png|Apoptosis signal pathway|400 px|thumb]] | ||
| Line 29: | Line 29: | ||
==Reference== | ==Reference== | ||
| + | <references/> | ||
1. RAPHAEL E. POLLOCK. ''Enhanced MDM2 oncoprotein expression in soft tissue sarcoma: several possible regulatory mechanisms''.''Sarcoma''. 1997. 1,23-29. | 1. RAPHAEL E. POLLOCK. ''Enhanced MDM2 oncoprotein expression in soft tissue sarcoma: several possible regulatory mechanisms''.''Sarcoma''. 1997. 1,23-29. | ||
| - | + | ||
[[Category: Xenopus laevis]] | [[Category: Xenopus laevis]] | ||
[[Category: P53 Tumor Suppressor]] | [[Category: P53 Tumor Suppressor]] | ||
Revision as of 03:27, 14 September 2012
Template:STRUCTURE 1ycqContents |
Role in Cancer
MDM2 is an active inhibitor of the p53 anti-tumorgenesis protein. When MDM2 is atached p53 is unable to release transcription factors that code for apoptosis or halting of the cell cycle. The full cell signaling pathway is very complex, but MDM2 overproduction has been shown to be connected to cancer activity.[1] Genetic amplification of MDM2 has been shown in breast, glioma, pancreaticadenocarcinoma, leucemias, Hodgkin's and nonhodgkin's lymphomas and other various cancers. [1] This broad observation outlines the importance of determining MDM2's role in tumer-genesis and how the process can be treated.
Experimental Research
Early work in the field of MDM2 overproduction in tumor cells was done by Raphael E. Pollok et. al. This research consisted of tests done with cancerous tumor tissue and analogous tissue. DNA amplification, mRNA creation, and protein creation were all areas of interest in determining what mechanism was being utilized for the overexertion of MDM2. The amplification of the DNA would show a DNA based increase in coding for the protein. An excess of mRNA would show the increase in protein was due to increased transcription factors for MDM2. The last possibility would be that the overproduction of MDM2 was an error in translation and only protein production was to blame.
These factors were tested using Southern, Northern, and Western plots to analyze DNA, mRNA, and protein production respectively. These tests use targeting antigens to test for the presence of desired proteins. The results of these tests reveled the genetic amplification of the DNA was to blame for the increased of MDM2. Also evidence that p53 protein mutations affect MDM2 were found. These results are of great use to focus treatment possibilities and further research.[1]
3D structures of MDM2
1ycq, 1ycr – fMDM2 + p53 transactivation domain – frog
1rv1, 1t4e, 3lbk – hMDM2 (mutant) p53-binding domain + inhibitor – human
3jzk, 3lbl, 3tu1 - hMDM2 p53-binding domain + inhibitor
1t4f - hMDM2 (mutant) p53-binding domain + p53 peptide
2axi, 2gv2, 3eqs, 3g03, 3iux, 3lnj, 3lnz, 3iwy - hMDM2 p53-binding domain + peptide
1z1m – hMDM2 N terminal – NMR
2c6a, 2c6b – hMDM2 C4 Zinc-finger domain – NMR
2hdp - hMDM2 C4 ring-finger domain – NMR
1ttv - fMDM2 (mutant) p53-binding domain + inhibitor – NMR
2vje, 2vjf – hMDM2 residues 383-446 + hMDM4 residues 428-490
Additional Resources
For additional information, see: Cancer
Reference
- ↑ Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996 Nov 8;274(5289):948-53. PMID:8875929
1. RAPHAEL E. POLLOCK. Enhanced MDM2 oncoprotein expression in soft tissue sarcoma: several possible regulatory mechanisms.Sarcoma. 1997. 1,23-29.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Matt Routh, David Canner, Joel L. Sussman, Yousuf Bahrami, Alexander Berchansky