We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3kli
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
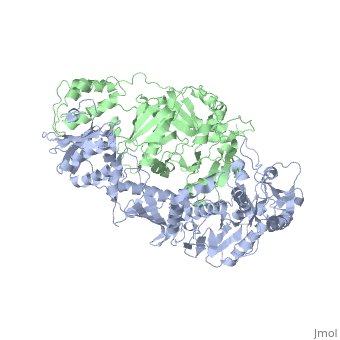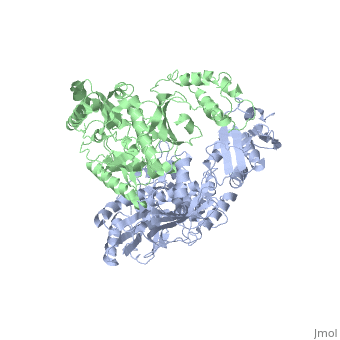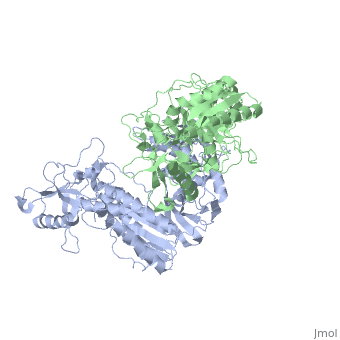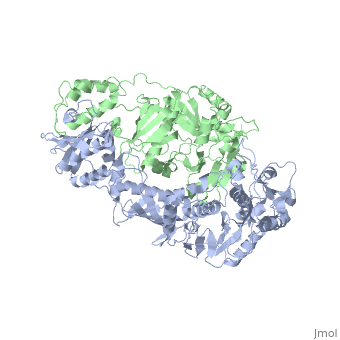
| - | [[ | + | ==Crystal structure of unliganded AZT-resistant HIV-1 Reverse Transcriptase== |
| + | <StructureSection load='3kli' size='340' side='right' caption='[[3kli]], [[Resolution|resolution]] 2.65Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[3kli]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Human_immunodeficiency_virus_1 Human immunodeficiency virus 1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3KLI OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3KLI FirstGlance]. <br> | ||
| + | </td></tr><tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3kle|3kle]], [[3klf|3klf]], [[3klg|3klg]], [[3klh|3klh]]</td></tr> | ||
| + | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">gag-pol ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=11676 Human immunodeficiency virus 1])</td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3kli FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3kli OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3kli RCSB], [http://www.ebi.ac.uk/pdbsum/3kli PDBsum]</span></td></tr> | ||
| + | </table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/kl/3kli_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Human immunodeficiency virus (HIV-1) develops resistance to 3'-azido-2',3'-deoxythymidine (AZT, zidovudine) by acquiring mutations in reverse transcriptase that enhance the ATP-mediated excision of AZT monophosphate from the 3' end of the primer. The excision reaction occurs at the dNTP-binding site, uses ATP as a pyrophosphate donor, unblocks the primer terminus and allows reverse transcriptase to continue viral DNA synthesis. The excision product is AZT adenosine dinucleoside tetraphosphate (AZTppppA). We determined five crystal structures: wild-type reverse transcriptase-double-stranded DNA (RT-dsDNA)-AZTppppA; AZT-resistant (AZTr; M41L D67N K70R T215Y K219Q) RT-dsDNA-AZTppppA; AZTr RT-dsDNA terminated with AZT at dNTP- and primer-binding sites; and AZTr apo reverse transcriptase. The AMP part of AZTppppA bound differently to wild-type and AZTr reverse transcriptases, whereas the AZT triphosphate part bound the two enzymes similarly. Thus, the resistance mutations create a high-affinity ATP-binding site. The structure of the site provides an opportunity to design inhibitors of AZT-monophosphate excision. | ||
| - | + | Structural basis of HIV-1 resistance to AZT by excision.,Tu X, Das K, Han Q, Bauman JD, Clark AD Jr, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E Nat Struct Mol Biol. 2010 Oct;17(10):1202-9. Epub 2010 Sep 19. PMID:20852643<ref>PMID:20852643</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==See Also== | ==See Also== | ||
*[[AZT-resistant HIV-1 reverse transcriptase|AZT-resistant HIV-1 reverse transcriptase]] | *[[AZT-resistant HIV-1 reverse transcriptase|AZT-resistant HIV-1 reverse transcriptase]] | ||
*[[Reverse transcriptase|Reverse transcriptase]] | *[[Reverse transcriptase|Reverse transcriptase]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Human immunodeficiency virus 1]] | [[Category: Human immunodeficiency virus 1]] | ||
| - | [[Category: Arnold, E | + | [[Category: Arnold, E]] |
| - | [[Category: Sarafianos, S G | + | [[Category: Sarafianos, S G]] |
| - | [[Category: Tu, X | + | [[Category: Tu, X]] |
[[Category: Aid]] | [[Category: Aid]] | ||
[[Category: Azt]] | [[Category: Azt]] | ||
Revision as of 09:34, 9 December 2014
Crystal structure of unliganded AZT-resistant HIV-1 Reverse Transcriptase
| |||||||||||
Categories: Human immunodeficiency virus 1 | Arnold, E | Sarafianos, S G | Tu, X | Aid | Azt | Azt resistance | Azt resistance mechanism | Azt resistance mutation | Dna polymerase | Dna recombination | Hetero dimer | Hiv | Multifunctional enzyme | Nrti | Nucleoside inhibitor | P51/p66 | Reverse transcriptase | Rna-directed dna polymerase | Transferase