We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Fragment-Based Drug Discovery
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='' size='500' side='right' caption='Bcl-xl in complex with ABT-737 (PDB entry [[2yxj]])' scene='Sandbox_reserved_394/Bcl-xl_abt-737_complex/2'> | <StructureSection load='' size='500' side='right' caption='Bcl-xl in complex with ABT-737 (PDB entry [[2yxj]])' scene='Sandbox_reserved_394/Bcl-xl_abt-737_complex/2'> | ||
Traditionally, new drugs are developed by either making small changes to existing drugs or by individually testing thousands of compounds. Both of these methods require many hours of laborious chemical synthesis. However, new techniques are being applied in the drug industry which decrease the cost and time required to discover and develop new drugs. | Traditionally, new drugs are developed by either making small changes to existing drugs or by individually testing thousands of compounds. Both of these methods require many hours of laborious chemical synthesis. However, new techniques are being applied in the drug industry which decrease the cost and time required to discover and develop new drugs. | ||
| + | |||
__TOC__ | __TOC__ | ||
| + | |||
=== Fragment-Based Drug Discovery === | === Fragment-Based Drug Discovery === | ||
---- | ---- | ||
| Line 10: | Line 12: | ||
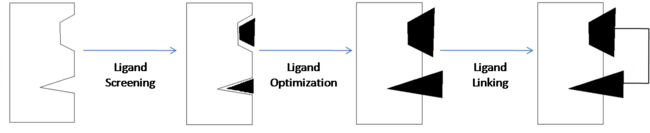
[[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery]] | [[Image:SAR by NMR Illustrated.png | thumb | center | 650px | Fragment-Based Drug Discovery]] | ||
| - | The development of <scene name='Sandbox_reserved_394/Abt-737/1'>ABT-737</scene> using | + | |
| + | The development of <scene name='Sandbox_reserved_394/Abt-737/1'>ABT-737</scene> using SAR by NMR is a classic example of FBDD. (Throughout this discussion ABT-737 will be used to illustrate the FBDD process.) This compound has been shown to effectively inhibit the over-expression of <scene name='Sandbox_reserved_394/Bcl-xl/1'>Bcl-xl</scene> which is a protein that is commonly observed to be over-expressed in many types of cancers.<ref name="Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579">Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579</ref> It acts an inhibitor of apoptosis and may also contribute to chemotherapy resistance. Bcl-xl inhibition by ABT-737 therefore, allows apoptosis to occur and helps to prevent chemo-resistance. | ||
{| class="wikitable collapsible collapsed" | {| class="wikitable collapsible collapsed" | ||
! scope="col" width="5000px" | SAR by NMR | ! scope="col" width="5000px" | SAR by NMR | ||
| Line 20: | Line 23: | ||
The first step of FBDD is to expose the potential drug target to a large number of small molecular fragments. This is usually done with a method known as high-throughput screening. [http://www.http://en.wikipedia.org/wiki/High-throughput_screening High-throughput screening] (HTS) is the process of using robotics to perform a large number of chemical tests. HTS is used to quickly identify fragments that have affinity for the target which are then analyzed to understand why they have affinity. | The first step of FBDD is to expose the potential drug target to a large number of small molecular fragments. This is usually done with a method known as high-throughput screening. [http://www.http://en.wikipedia.org/wiki/High-throughput_screening High-throughput screening] (HTS) is the process of using robotics to perform a large number of chemical tests. HTS is used to quickly identify fragments that have affinity for the target which are then analyzed to understand why they have affinity. | ||
| + | |||
| + | ===== ABT-737: ligand screening ===== | ||
| + | |||
| + | <scene name='Sandbox_reserved_394/Compound_1/1'>Two fragments</scene> were found to have moderate affinity for Bcl-xl. <scene name='Sandbox_reserved_394/Compound_1/2'>Compound 1</scene> is a fluorobiphenylcarboxylic acid. It occupies <scene name='Sandbox_reserved_394/Binding_site_1/1'>binding site 1</scene> of Bcl-xl. The fluorobiphenyl portion of compound 1 is very hydrophobic. Therefore, Bcl-xl forms a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the fluorobiphenyl system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxyilic acid portion of compound 1 binds near Gly 142</scene> of Bcl-xl. | ||
| + | |||
| + | <scene name='Sandbox_reserved_394/Compound_1/3'>Compound 2</scene> is a napthalene-based alcohol which occupies <scene name='Sandbox_reserved_394/Binding_site_2/1'>binding site 2</scene>. | ||
==== Ligand Optimization ==== | ==== Ligand Optimization ==== | ||
Once the fragments have been identified, they are then modified to increase their binding affinity. | Once the fragments have been identified, they are then modified to increase their binding affinity. | ||
| + | |||
| + | ===== ABT-737: ligand optimization ===== | ||
Applying these 3-D structures to the drug design process involves using either structure-based drug design (SBDD) or ligand-based drug design (LBDD). | Applying these 3-D structures to the drug design process involves using either structure-based drug design (SBDD) or ligand-based drug design (LBDD). | ||
Revision as of 02:46, 31 October 2012
Drug Design: Fragment-Based Drug Discovery
| |||||||||||
References
- ↑ Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579
- ↑ Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.
- ↑ Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf