We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
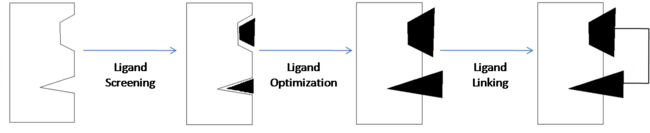
Fragment-Based Drug Discovery
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
<scene name='Sandbox_reserved_394/Compound_1/1'>Two fragments</scene> were found to have moderate affinity for Bcl-xl. <scene name='Sandbox_reserved_394/Compound_1/2'>Compound 1</scene> is a fluorobiphenylcarboxylic acid. It occupies <scene name='Sandbox_reserved_394/Binding_site_1/1'>binding site 1</scene> of Bcl-xl. The fluorobiphenyl portion of compound 1 is very hydrophobic. Therefore, Bcl-xl forms a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the fluorobiphenyl system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxyilic acid portion of compound 1 binds near Gly 142</scene> of Bcl-xl. | <scene name='Sandbox_reserved_394/Compound_1/1'>Two fragments</scene> were found to have moderate affinity for Bcl-xl. <scene name='Sandbox_reserved_394/Compound_1/2'>Compound 1</scene> is a fluorobiphenylcarboxylic acid. It occupies <scene name='Sandbox_reserved_394/Binding_site_1/1'>binding site 1</scene> of Bcl-xl. The fluorobiphenyl portion of compound 1 is very hydrophobic. Therefore, Bcl-xl forms a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the fluorobiphenyl system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxyilic acid portion of compound 1 binds near Gly 142</scene> of Bcl-xl. | ||
| - | <scene name='Sandbox_reserved_394/Compound_1/3'>Compound 2</scene> is a napthalene-based alcohol which occupies <scene name='Sandbox_reserved_394/Binding_site_2/1'>binding site 2</scene>. | + | <scene name='Sandbox_reserved_394/Compound_1/3'>Compound 2</scene> is a napthalene-based alcohol which occupies <scene name='Sandbox_reserved_394/Binding_site_2/1'>binding site 2</scene>. This particular fragment also is involved with hydrophobic interactions with Bcl-xl, although they are not as strong as in the case of compound 1. |
==== Ligand Optimization ==== | ==== Ligand Optimization ==== | ||
| - | Once the fragments have been identified, they are then modified to increase their binding affinity. | + | Once the fragments have been identified, they are then modified to increase their binding affinity. These modifications can include atom substitutions, the addition of substituents, or even the replacement of the entire fragment. Knowing and having an understanding of the structure of the biological target is useful in optimizing the fragments. The nature of the binding site is what determines how a ligand will bind (as in the case of the hydrophobic pocket formed around compound 1). This approach to designing drugs is referred to as structure-based drug design. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| class="wikitable collapsible collapsed" | {| class="wikitable collapsible collapsed" | ||
! scope="col" width="5000px" | Structure-Based Drug Design | ! scope="col" width="5000px" | Structure-Based Drug Design | ||
| Line 42: | Line 38: | ||
| scope="col" width="5000px" | Structure-based drug design is utilized when the 3-D structure of a protein, or other drug target, is used to predict drug candidates. A visual representation of the structure allows developers to pinpoint binding sites and more effectively design a drug that will have high affinity for the target. | | scope="col" width="5000px" | Structure-based drug design is utilized when the 3-D structure of a protein, or other drug target, is used to predict drug candidates. A visual representation of the structure allows developers to pinpoint binding sites and more effectively design a drug that will have high affinity for the target. | ||
|} | |} | ||
| + | |||
| + | ===== ABT-737: ligand optimization ===== | ||
| + | |||
| + | Compounds 1 & 2 exhibited very poor binding affinity for Bcl-xl. The optimization of these two compounds resulted in <scene name='Sandbox_reserved_394/Compound_2/1'>Compound 2</scene>. In order to improve the binding affinity, the carboxylic acid of compound 1 was substituted with an acyl sulfonamide to capitalize on the hydrophilic interaction with the protein. This <scene name='Sandbox_reserved_394/Compound_2/2'>acylsulfonamide portion forms a hydrogen bond with Gly 142</scene> thereby increasing the affinity for Bcl-xl. | ||
| + | |||
| + | Applying these 3-D structures to the drug design process involves using either structure-based drug design (SBDD) or ligand-based drug design (LBDD). | ||
| + | |||
{| class="wikitable collapsible collapsed" | {| class="wikitable collapsible collapsed" | ||
! scope="col" width="5000px" | Ligand-Based Drug Design | ! scope="col" width="5000px" | Ligand-Based Drug Design | ||
Revision as of 03:08, 31 October 2012
Drug Design: Fragment-Based Drug Discovery
| |||||||||||
References
- ↑ Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579
- ↑ Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.
- ↑ Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf