We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Fragment-Based Drug Discovery
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
! scope="col" width="5000px" | SAR by NMR | ! scope="col" width="5000px" | SAR by NMR | ||
|- | |- | ||
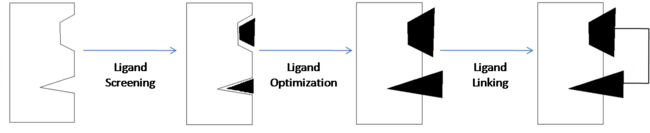
| - | | scope="col" width="5000px" | Structure-activity relationship (SAR) by NMR is one tool that is commonly used to design and develop new drugs. This is the process "in which small organic molecules that bind to proximal subsites of a protein are identified, optimized, and linked together to produce high-affinity ligands."<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref> | + | | scope="col" width="5000px" | Structure-activity relationship (SAR) by NMR is one tool that is commonly used to design and develop new drugs. This is the process "in which small organic molecules that bind to proximal subsites of a protein are identified, optimized, and linked together to produce high-affinity ligands."<ref name="Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.">Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.</ref> In other words, NMR is used to identify the components responsible for binding and analyze the relationship between the ligand and the biological target. |
|} | |} | ||
| Line 43: | Line 43: | ||
Compounds 1 & 2 exhibited very poor binding affinity for Bcl-xl. The optimization of these two compounds resulted in <scene name='Sandbox_reserved_394/Compound_2/1'>Compound 3</scene>. In order to improve the binding affinity, the carboxylic acid of compound 1 was substituted with an acyl sulfonamide to capitalize on the hydrophilic interaction with the protein. This <scene name='Sandbox_reserved_394/Compound_2/2'>acylsulfonamide portion forms a hydrogen bond with Gly 142</scene> thereby increasing the affinity for Bcl-xl. The substitution of the sulfonamide actually allows the acidic proton to get closer to Gly 142 than it could in the carboxylic acid, which is why it is able to bind stronger to the amino acid. | Compounds 1 & 2 exhibited very poor binding affinity for Bcl-xl. The optimization of these two compounds resulted in <scene name='Sandbox_reserved_394/Compound_2/1'>Compound 3</scene>. In order to improve the binding affinity, the carboxylic acid of compound 1 was substituted with an acyl sulfonamide to capitalize on the hydrophilic interaction with the protein. This <scene name='Sandbox_reserved_394/Compound_2/2'>acylsulfonamide portion forms a hydrogen bond with Gly 142</scene> thereby increasing the affinity for Bcl-xl. The substitution of the sulfonamide actually allows the acidic proton to get closer to Gly 142 than it could in the carboxylic acid, which is why it is able to bind stronger to the amino acid. | ||
| - | Compound 2 was important in identifying the hydrophobicity of binding site 2 but was substituted with a <scene name='Sandbox_reserved_394/Nitro_thio_phenyl_sub/1'>3-nitro-4-(2-phenylthioethyl)aminophenyl group</scene>. This substitution | + | Compound 2 was important in identifying the hydrophobicity of binding site 2 but was substituted with a <scene name='Sandbox_reserved_394/Nitro_thio_phenyl_sub/1'>3-nitro-4-(2-phenylthioethyl)aminophenyl group</scene>. This substitution more efficiently binds to site 2 through π-π stacking. The idea of using a known ligand to develop another ligand, and eventually drugs, is known as ligand-based drug design. |
| - | + | ||
| - | + | ||
{| class="wikitable collapsible collapsed" | {| class="wikitable collapsible collapsed" | ||
| Line 51: | Line 49: | ||
|- | |- | ||
| scope="col" width="5000px" | "Ligand-based drug design (LBDD) techniques are applied when the structure of the receptor is unknown but when a series of compounds or ligands have been identified that show the biological activity of the interest."<ref name="Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf">Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf</ref> In other words, once it is known how a ligand binds to a protein or any other molecule, new ligands, and eventually drugs, can be designed to bind in a similar manner and get the desired effect. It involves modifying a known ligand to develop another ligand with a higher binding affinity for the target. | | scope="col" width="5000px" | "Ligand-based drug design (LBDD) techniques are applied when the structure of the receptor is unknown but when a series of compounds or ligands have been identified that show the biological activity of the interest."<ref name="Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf">Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf</ref> In other words, once it is known how a ligand binds to a protein or any other molecule, new ligands, and eventually drugs, can be designed to bind in a similar manner and get the desired effect. It involves modifying a known ligand to develop another ligand with a higher binding affinity for the target. | ||
| - | |} | + | |} |
| + | |||
| + | Compound 3 required further optimization because the binding affinity for Bcl-xl is greatly reduced in the presence of human serum albumin (HSA). In order to decrease HSA affinity, and therefore increase Bcl-xl affinity, SAR by NMR was used to modify compound 3 by eliminating key binding groups of the compound to HSA without affecting Bcl-xl affinity. | ||
| + | |||
| + | {| class="wikitable collapsible collapsed" | ||
| + | ! scope="col" width="5000px" | Modifying compound 2 to reduce HSA affinity | ||
| + | |- | ||
| + | | scope="col" width="5000px" | Compound 2 has high affinity for Bcl-xl but has an even higher affinity for HSA. For this reason, when HSA is present, compound 2 and similar ligands are more likely to bind to HSA thereby decreasing the amount that can bind with Bcl-xl. In order to decrease the affinity for HSA while maintaining affinity for Bcl-xl, SAR by NMR was used to compare compound 2 with a <scene name='Sandbox_reserved_394/Compound_3/1'>thioethylamino-2,4-dimethylphenyl analogue</scene>, which also has high affinity for HSA. It was found that two hydrophobic portions of compound 2 had very strong hydrophobic interactions with HSA. Therefore, these portions were modified with polar substituents to decrease HSA affinity. To decrease hydrophobicity, the fluorobiphenyl system was substituted with a piperazine ring and a 2-dimethylaminoethyl group was added to the thioethylamino linkage group. | ||
| + | |} | ||
| + | |||
| + | ==== Ligand Linking ==== | ||
| + | |||
| + | |||
| + | |||
| + | Applying these 3-D structures to the drug design process involves using either structure-based drug design (SBDD) or ligand-based drug design (LBDD). | ||
==== ABT-737 ==== | ==== ABT-737 ==== | ||
| Line 63: | Line 75: | ||
<scene name='Sandbox_reserved_394/Compound_1/2'>Compound 1</scene> is a 4'-fluoro-biphenyl-4-carboxylic acid. SAR by NMR was used to identify the interactions that this compound forms with Bcl-xl. The fluorobiphenyl system is hydrophobic and its interactions form a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the fluorobiphenyl system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxyilic acid portion binds near Gly 142</scene> of Bcl-xl. The carboxylic acid is later substituted with an acyl sulfonamide (shown in compound 2) which provides increased affinity. | <scene name='Sandbox_reserved_394/Compound_1/2'>Compound 1</scene> is a 4'-fluoro-biphenyl-4-carboxylic acid. SAR by NMR was used to identify the interactions that this compound forms with Bcl-xl. The fluorobiphenyl system is hydrophobic and its interactions form a <scene name='Sandbox_reserved_394/Compound_1/4'>"hydrophobic pocket"</scene> around the fluorobiphenyl system. The <scene name='Sandbox_reserved_394/Compound_1/5'>carboxyilic acid portion binds near Gly 142</scene> of Bcl-xl. The carboxylic acid is later substituted with an acyl sulfonamide (shown in compound 2) which provides increased affinity. | ||
| - | <scene name='Sandbox_reserved_394/Compound_2/1'>Compound 2</scene> binds with high affinity to Bcl-xl. The <scene name='Sandbox_reserved_394/Compound_2/2'>acylsulfonamide portion forms a hydrogen bond with Gly 142</scene>. The substitution of this sulfonamide for the carboxylic acid from compound 1 allows compound 2 to form a much stronger bond with Bcl-xl by bringing the shared acidic proton in closer proximity to GLY 142. The | + | <scene name='Sandbox_reserved_394/Compound_2/1'>Compound 2</scene> binds with high affinity to Bcl-xl. The <scene name='Sandbox_reserved_394/Compound_2/2'>acylsulfonamide portion forms a hydrogen bond with Gly 142</scene>. The substitution of this sulfonamide for the carboxylic acid from compound 1 allows compound 2 to form a much stronger bond with Bcl-xl by bringing the shared acidic proton in closer proximity to GLY 142. The |
| + | |||
| - | {| class="wikitable collapsible collapsed" | ||
| - | ! scope="col" width="5000px" | Modifying compound 2 to reduce HSA affinity | ||
| - | |- | ||
| - | | scope="col" width="5000px" | Compound 2 has high affinity for Bcl-xl but has an even higher affinity for HSA. For this reason, when HSA is present, compound 2 and similar ligands are more likely to bind to HSA thereby decreasing the amount that can bind with Bcl-xl. In order to decrease the affinity for HSA while maintaining affinity for Bcl-xl, SAR by NMR was used to compare compound 2 with a <scene name='Sandbox_reserved_394/Compound_3/1'>thioethylamino-2,4-dimethylphenyl analogue</scene>, which also has high affinity for HSA. It was found that two hydrophobic portions of compound 2 had very strong hydrophobic interactions with HSA. Therefore, these portions were modified with polar substituents to decrease HSA affinity. To decrease hydrophobicity, the fluorobiphenyl system was substituted with a piperazine ring and a 2-dimethylaminoethyl group was added to the thioethylamino linkage group. | ||
| - | |} | ||
Once the components responsible for binding are identified, they can be modified, as in the case of compound 1 where the carboxylic acid was substituted with an acyl sulfonamide, and then they are linked together to create a compound with optimal binding affinity. | Once the components responsible for binding are identified, they can be modified, as in the case of compound 1 where the carboxylic acid was substituted with an acyl sulfonamide, and then they are linked together to create a compound with optimal binding affinity. | ||
Revision as of 16:19, 31 October 2012
Drug Design: Fragment-Based Drug Discovery
| |||||||||||
References
- ↑ Oltersdorf T., Elmore S. W., Shoemaker A. R. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Vol 435|2 June 2005|doi:10.1038/nature03579
- ↑ Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science; Nov 29, 1996; 274, 5292; ProQuest Central pg. 1531.
- ↑ Pandit D. LIGAND-BASED DRUG DESIGN: I. CONFORMATIONAL STUDIES OF GBR 12909 ANALOGS AS COCAINE ANTAGONISTS; II. 3D-QSAR STUDIES OF SALVINORIN A ANALOGS AS εΑΡΡΑ OPIOID AGONISTS. http://archives.njit.edu/vol01/etd/2000s/2007/njit-etd2007-051/njit-etd2007-051.pdf