We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Tutorial:Basic Chemistry Topics
From Proteopedia
(Difference between revisions)
| Line 45: | Line 45: | ||
*Hydrogen Bonds, the weakest of bonds, are attractive interactions between an electronegative atom and hydrogen. Electronegative atoms are atoms that have high electron density. They are strong atoms that pull electrons towards them from weaker/low electron density atoms, such as hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. Water is the most common example of hydrogen bonding. The water molecule chemical formula is H2O. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons and allowing the formation of a water droplet. The electronegative atoms allow for the droplet to be held together instead of spreading. In this representation the hydrogen bonds are represented as yellow-dashed lines. The hydrogen bonds are important to the stability of the secondary structures. | *Hydrogen Bonds, the weakest of bonds, are attractive interactions between an electronegative atom and hydrogen. Electronegative atoms are atoms that have high electron density. They are strong atoms that pull electrons towards them from weaker/low electron density atoms, such as hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. Water is the most common example of hydrogen bonding. The water molecule chemical formula is H2O. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons and allowing the formation of a water droplet. The electronegative atoms allow for the droplet to be held together instead of spreading. In this representation the hydrogen bonds are represented as yellow-dashed lines. The hydrogen bonds are important to the stability of the secondary structures. | ||
| - | + | <scene name='Tutorial:Basic_Chemistry_Topics/Hydrogen_bonds/2'>Hydrogen Bonds</scene> | |
='''Secondary Structures'''= | ='''Secondary Structures'''= | ||
| - | Secondary structures are alpha helices and beta sheets, which help contribute to the stability of the molecule. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately four alpha helices and two beta strands when presented as a monomer. Since this structure is represented as a dimer you actually have eight alpha helices and four beta sheets. The concept of a dimer is explained in the "Ligands" section later on in the tutorial. Alpha helices rotate in a clockwise manner and are also oriented in a parallel formation. The parallel alpha helices are held together by hydrogen bond, which we discussed earlier. Beta sheets are often anti-parallel. The structure of the alpha and beta sheets in Tuberculosis/CoA/Tobramycin structure represents the GNAT fold. The folding of a protein is what gives the function. When a change occurs in the folding, a change in the function also takes place. The GNAT fold described in the study has a function of acetylation, which is the addition of an acyl group. The chemical formula of an acetyl group is COCH3. It is important to note that the discovery of the GNAT fold lead to the understanding of the major function. | + | *Secondary structures are alpha helices and beta sheets, which help contribute to the stability of the molecule. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately four alpha helices and two beta strands when presented as a monomer. Since this structure is represented as a dimer you actually have eight alpha helices and four beta sheets. The concept of a dimer is explained in the "Ligands" section later on in the tutorial. Alpha helices rotate in a clockwise manner and are also oriented in a parallel formation. The parallel alpha helices are held together by hydrogen bond, which we discussed earlier. Beta sheets are often anti-parallel. The structure of the alpha and beta sheets in Tuberculosis/CoA/Tobramycin structure represents the GNAT fold. The folding of a protein is what gives the function. When a change occurs in the folding, a change in the function also takes place. The GNAT fold described in the study has a function of acetylation, which is the addition of an acyl group. The chemical formula of an acetyl group is COCH3. It is important to note that the discovery of the GNAT fold lead to the understanding of the major function. |
| - | <scene name='Tutorial:Basic_Chemistry_Topics/ | + | <scene name='Tutorial:Basic_Chemistry_Topics/Alpha_beta_2ndstructures/1'>Alpha and Beta Strands</scene> |
='''Active Site'''= | ='''Active Site'''= | ||
| - | The active site of a molecule can be described as a pocket where an interaction between substrates causes a physiological effect by causing a change in conformation. The conformation is referring to the orientation of the molecules involved in the structure. The conformation change can inhibit or activate the physiological effect. The active site is where the ligand is going to bind. (Ligands are discussed in detail later on in the “Ligands” section) The active site can either be inhibited or activated by ligands. Referring back to our article, the active site is where the substrate, in this case tobramycin, binds to CoA and the mycobacterium to cause an antibacterial effect. It the study described, this is where the acetylation of the tobramycin should be occurring. The acetylation of tobramycin would cause the tobramycin to be inactive, hence inhibiting the active site. | + | *The active site of a molecule can be described as a pocket where an interaction between substrates causes a physiological effect by causing a change in conformation. The conformation is referring to the orientation of the molecules involved in the structure. The conformation change can inhibit or activate the physiological effect. The active site is where the ligand is going to bind. (Ligands are discussed in detail later on in the “Ligands” section) The active site can either be inhibited or activated by ligands. Referring back to our article, the active site is where the substrate, in this case tobramycin, binds to CoA and the mycobacterium to cause an antibacterial effect. It the study described, this is where the acetylation of the tobramycin should be occurring. The acetylation of tobramycin would cause the tobramycin to be inactive, hence inhibiting the active site. |
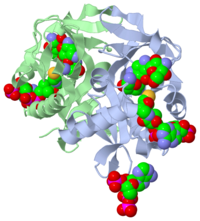
='''Ligand'''= | ='''Ligand'''= | ||
| - | Ligands are molecules or complexes that are within the secondary structures that orient in such a way to contribute the function of the complex as a whole. Ligands can have binding sites on receptors, and when bound can trigger a physiological response. A ligand can be a competitive agonist, allosteric agonist, competitive antagonist, or an allosteric antagonist. An agonist is a ligand that causes a physiological response, activating the active site. An antagonist is a ligand that inhibits a physiological response, not allowing the active site to be activated. | + | *Ligands are molecules or complexes that are within the secondary structures that orient in such a way to contribute the function of the complex as a whole. Ligands can have binding sites on receptors, and when bound can trigger a physiological response. A ligand can be a competitive agonist, allosteric agonist, competitive antagonist, or an allosteric antagonist. An agonist is a ligand that causes a physiological response, activating the active site. An antagonist is a ligand that inhibits a physiological response, not allowing the active site to be activated. |
| - | A ligand is competitive when it is binding to the same site as the physiological activator; hence it is competing for the same site. When a ligand binds to an allosteric site, the ligand is binding to the same receptor but it is not binding to the active site. The ligands present in the complex used by the research article are coenzyme A, Tobramycin and Phosphate-Adenosine-5'-Diphosphate. | + | *A ligand is competitive when it is binding to the same site as the physiological activator; hence it is competing for the same site. When a ligand binds to an allosteric site, the ligand is binding to the same receptor but it is not binding to the active site. The ligands present in the complex used by the research article are coenzyme A, Tobramycin and Phosphate-Adenosine-5'-Diphosphate. |
==Coenzyme A== | ==Coenzyme A== | ||
| - | Coenzyme (CoA) is a coenzyme that synthesizes and oxidizes fatty acids. This process is essential for the utilization of fatty acids. Coenzyme A is used as a substrate in the citric acid cycle. The citric acid cycle is also known as the Krebs cycle or tricarboxylic acid cycle (TCA). This process is important to the production of ATP, which is an energy source used by the body. | + | *Coenzyme (CoA) is a coenzyme that synthesizes and oxidizes fatty acids. This process is essential for the utilization of fatty acids. Coenzyme A is used as a substrate in the citric acid cycle. The citric acid cycle is also known as the Krebs cycle or tricarboxylic acid cycle (TCA). This process is important to the production of ATP, which is an energy source used by the body. |
| - | The Protein’s in this molecule are represented as a dimer. A dimer is a chemical structure formed from two identical subunits. Some molecules are present as a dimer because it is more stable then the monomer. The dimer is constructed by connecting two subunits along their axis. | + | *The Protein’s in this molecule are represented as a dimer. A dimer is a chemical structure formed from two identical subunits. Some molecules are present as a dimer because it is more stable then the monomer. The dimer is constructed by connecting two subunits along their axis. |
==Tobramycin== | ==Tobramycin== | ||
| - | Tobramycin is an antibiotic part of the aminoglycoside family. Aminoglycosides produce antibacterial effects by inhibiting protein synthesis and compromising the cell wall structure. By inhibiting the protein synthesis of the bacteria, it does not allow the bacteria to replicate. The cell wall is an important structure to bacteria because it provides the structure and stability to the bacteria. By disrupting the cell wall, we are removing the stability of the bacteria and ultimately casing bacteria death. | + | *Tobramycin is an antibiotic part of the aminoglycoside family. Aminoglycosides produce antibacterial effects by inhibiting protein synthesis and compromising the cell wall structure. By inhibiting the protein synthesis of the bacteria, it does not allow the bacteria to replicate. The cell wall is an important structure to bacteria because it provides the structure and stability to the bacteria. By disrupting the cell wall, we are removing the stability of the bacteria and ultimately casing bacteria death. |
Tobramycin targets a variety of bacteria, particularly gram(-) species. Just like all drugs there are side effects associated with tobramycin. Some of the more common side effects are ototoxicity and nephrotoxicity. Ototoxic is hearing loss and nephrotoxic is causing kidney damage. The kidney damage is due to Tobramycin reabsorption through the renal tubules. This basically means that tobramycin may be toxic to the kidneys. The toxicity is caused by the contact-time in the renal tubules where the drug is located. | Tobramycin targets a variety of bacteria, particularly gram(-) species. Just like all drugs there are side effects associated with tobramycin. Some of the more common side effects are ototoxicity and nephrotoxicity. Ototoxic is hearing loss and nephrotoxic is causing kidney damage. The kidney damage is due to Tobramycin reabsorption through the renal tubules. This basically means that tobramycin may be toxic to the kidneys. The toxicity is caused by the contact-time in the renal tubules where the drug is located. | ||
Revision as of 18:16, 31 October 2012
| |||||||||||