Clegg TBP sandbox
From Proteopedia
| Line 3: | Line 3: | ||
{{STRUCTURE_1cdw| PDB=1cdw | SCENE= }} | {{STRUCTURE_1cdw| PDB=1cdw | SCENE= }} | ||
| - | === | + | ===TATA Binding Protein Overview=== |
| - | + | ||
| - | + | ||
| - | + | ||
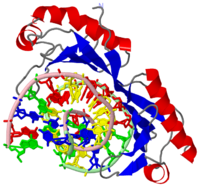
The TBP has two imperfect direct repeats that form a <scene name='Clegg_TBP_sandbox/Saddle/1'>saddle</scene> shaped structure forming a concave like DNA binding surface allowing for an induced-fit mechanism. The imperfect direct repeats differ by two residues. The first is a deletion of a resiude at the end of helix H1. The second is a proline in helix H2' that enables a <scene name='Clegg_TBP_sandbox/Proline/1'>bend</scene> that does not occur on H2. When the TBP binds to the TATA box a conformation change takes place and the DNA strand is <scene name='Clegg_TBP_sandbox/Bend/1'>bent</scene> 80 degrees. This conformational change promotes the recruitment of other general transcription factors. A total of seven <scene name='Clegg_TBP_sandbox/Lysines/1'>lysine</scene> residues interact with DNA backbone. Four of these residues Lys-204, Lys-214, Lys-295, and Lys 305 allow for the patial charge neutralization within the TATA box. Serveral arginines also interact with the DNA strand these residues include: <scene name='Clegg_TBP_sandbox/Lysines/3'>Arg-192</scene>, Arg-199, Arg-290. The TBP is crucual protien that recognizes the promoter region of DNA and allows for other general transcription factors to bind and form a complex which can eventually lead to transcripton. | The TBP has two imperfect direct repeats that form a <scene name='Clegg_TBP_sandbox/Saddle/1'>saddle</scene> shaped structure forming a concave like DNA binding surface allowing for an induced-fit mechanism. The imperfect direct repeats differ by two residues. The first is a deletion of a resiude at the end of helix H1. The second is a proline in helix H2' that enables a <scene name='Clegg_TBP_sandbox/Proline/1'>bend</scene> that does not occur on H2. When the TBP binds to the TATA box a conformation change takes place and the DNA strand is <scene name='Clegg_TBP_sandbox/Bend/1'>bent</scene> 80 degrees. This conformational change promotes the recruitment of other general transcription factors. A total of seven <scene name='Clegg_TBP_sandbox/Lysines/1'>lysine</scene> residues interact with DNA backbone. Four of these residues Lys-204, Lys-214, Lys-295, and Lys 305 allow for the patial charge neutralization within the TATA box. Serveral arginines also interact with the DNA strand these residues include: <scene name='Clegg_TBP_sandbox/Lysines/3'>Arg-192</scene>, Arg-199, Arg-290. The TBP is crucual protien that recognizes the promoter region of DNA and allows for other general transcription factors to bind and form a complex which can eventually lead to transcripton. | ||
Revision as of 03:49, 26 November 2012
| |||||||||
| 1cdw, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
TATA Binding Protein Overview
The TBP has two imperfect direct repeats that form a shaped structure forming a concave like DNA binding surface allowing for an induced-fit mechanism. The imperfect direct repeats differ by two residues. The first is a deletion of a resiude at the end of helix H1. The second is a proline in helix H2' that enables a that does not occur on H2. When the TBP binds to the TATA box a conformation change takes place and the DNA strand is 80 degrees. This conformational change promotes the recruitment of other general transcription factors. A total of seven residues interact with DNA backbone. Four of these residues Lys-204, Lys-214, Lys-295, and Lys 305 allow for the patial charge neutralization within the TATA box. Serveral arginines also interact with the DNA strand these residues include: , Arg-199, Arg-290. The TBP is crucual protien that recognizes the promoter region of DNA and allows for other general transcription factors to bind and form a complex which can eventually lead to transcripton.
About this Structure
1cdw is a 3 chain structure with sequence from Homo sapiens. The July 2005 RCSB PDB Molecule of the Month feature on TATA-Binding Protein by David S. Goodsell is 10.2210/rcsb_pdb/mom_2005_7. Full crystallographic information is available from OCA.
See Also
Reference
- Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4862-7. PMID:8643494
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000 Nov;1(5):411-5. PMID:11258480 doi:10.1093/embo-reports/kvd092
- Hicks JM, Hsu VL. The extended left-handed helix: a simple nucleic acid-binding motif. Proteins. 2004 May 1;55(2):330-8. PMID:15048824 doi:10.1002/prot.10630