Joe Granger Methionine Repressor: Escherichia coli
From Proteopedia
(→MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE) |
(→MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE) |
||
| Line 5: | Line 5: | ||
===MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE=== | ===MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE=== | ||
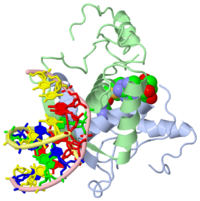
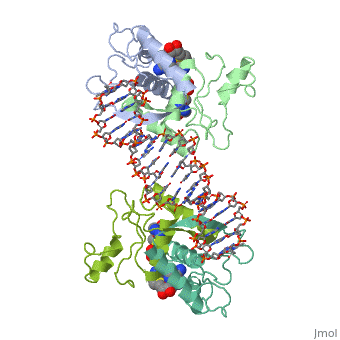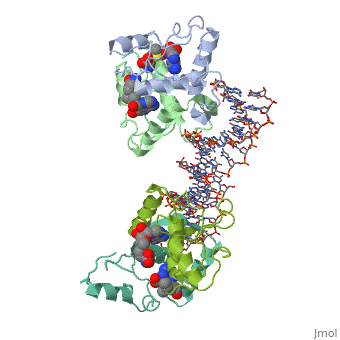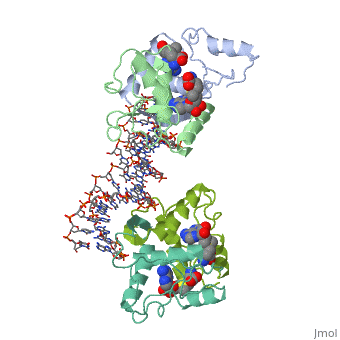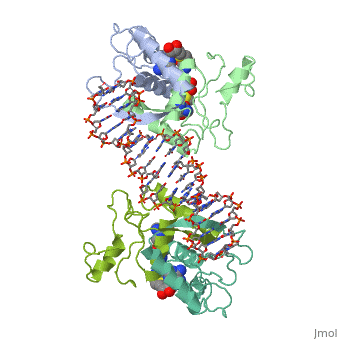
| - | {{ The Met Repressor Protein is involved with regulating the synthesis of methionine. This protein is a homodimer composed of 2 104 amino acid chains. The Met Repressor works to bind the DNA operators that activate the transcription of methionine producing genes. When this protein is bound to DNA, it prevents RNA polymerase'''II''' from being able to bind and promot transcription. It achieves this by taking the place of the activators on the operon. The crystal structure of the met repressor-operator complex shows two dimeric repressor molecules bound to adjacent sites 8 base pairs apart on an 18-base-pair DNA fragment. The <scene name='User:Wally_Novak/Suface_of_the_protein/6'>surface of the protein</scene> is bound tightly with The DNA operator in the major groove. What binds the protein to the DNA tightly is the two Beta sheets. While they are both from different monomers, they complex in an <scene name='User:Wally_Novak/Anti-parallel_h-bonding/1'> anti-parallel fashion</scene>. This complexed beta sheet not only connects the two monomers, but it is also involved with binding to the DNA <scene name='User:Wally_Novak/Met_repressor_concensious/1'> concensious sequence AGACGTCT </scene> . The beta sheets then can create the <scene name='User:Wally_Novak/Polar_bonding_to_dna/2'> polar conections</scene> necessary to attach to the concensious sequence. Sequence specificity to the concensious sequence is achieved by insertion of the antiparallel protein beta-ribbons into the major groove of the B-form DNA. There, the | + | {{ The Met Repressor Protein is involved with regulating the synthesis of methionine. This protein is a homodimer composed of 2 104 amino acid chains. The Met Repressor works to bind the DNA operators that activate the transcription of methionine producing genes. When this protein is bound to DNA, it prevents RNA polymerase'''II''' from being able to bind and promot transcription. It achieves this by taking the place of the activators on the operon. The crystal structure of the met repressor-operator complex shows two dimeric repressor molecules bound to adjacent sites 8 base pairs apart on an 18-base-pair DNA fragment. The <scene name='User:Wally_Novak/Suface_of_the_protein/6'>surface of the protein</scene> is bound tightly with The DNA operator in the major groove. What binds the protein to the DNA tightly is the two Beta sheets. While they are both from different monomers, they complex in an <scene name='User:Wally_Novak/Anti-parallel_h-bonding/1'> anti-parallel fashion</scene>. This complexed beta sheet not only connects the two monomers, but it is also involved with binding to the DNA <scene name='User:Wally_Novak/Met_repressor_concensious/1'> concensious sequence AGACGTCT </scene> . The beta sheets then can create the <scene name='User:Wally_Novak/Polar_bonding_to_dna/2'> polar conections</scene> necessary to attach to the concensious sequence. Sequence specificity to the concensious sequence is achieved by insertion of the antiparallel protein beta-ribbons into the major groove of the B-form DNA. There, the repressor recognizes sequence-dependent distortion or flexibility of the operator phosphate backbone, conferring specificity even for inaccessible base pairs. As shown by this mutant,() that has a mutation in its... Ultimately, the specific amino acid sequence and shape allows for it to bind to its consensus sequence }} |
==About this Structure== | ==About this Structure== | ||
Revision as of 03:14, 8 November 2012
| |||||||||
| 1cma, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE
{{ The Met Repressor Protein is involved with regulating the synthesis of methionine. This protein is a homodimer composed of 2 104 amino acid chains. The Met Repressor works to bind the DNA operators that activate the transcription of methionine producing genes. When this protein is bound to DNA, it prevents RNA polymeraseII from being able to bind and promot transcription. It achieves this by taking the place of the activators on the operon. The crystal structure of the met repressor-operator complex shows two dimeric repressor molecules bound to adjacent sites 8 base pairs apart on an 18-base-pair DNA fragment. The is bound tightly with The DNA operator in the major groove. What binds the protein to the DNA tightly is the two Beta sheets. While they are both from different monomers, they complex in an . This complexed beta sheet not only connects the two monomers, but it is also involved with binding to the DNA . The beta sheets then can create the necessary to attach to the concensious sequence. Sequence specificity to the concensious sequence is achieved by insertion of the antiparallel protein beta-ribbons into the major groove of the B-form DNA. There, the repressor recognizes sequence-dependent distortion or flexibility of the operator phosphate backbone, conferring specificity even for inaccessible base pairs. As shown by this mutant,() that has a mutation in its... Ultimately, the specific amino acid sequence and shape allows for it to bind to its consensus sequence }}
About this Structure
1cma is a 4 chain structure with sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
- Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387-93. PMID:1406951 doi:http://dx.doi.org/10.1038/359387a0
Santanu Maitra, and james S. Nowick. (2000) B-Sheet Interactions Between Proteins. University of Califonia Irvine. 15