Polymerase III homoenzyme beta subunit
From Proteopedia
(→Polymerase III Holoenzyme beta subunit) |
(→Polymerase III Holoenzyme beta subunit) |
||
| Line 1: | Line 1: | ||
== Polymerase III Holoenzyme beta subunit == | == Polymerase III Holoenzyme beta subunit == | ||
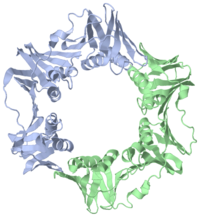
| + | [[Image:2pol.png|left|200px]] | ||
| + | |||
| + | {{STRUCTURE_2pol| PDB=2pol | SCENE= }} | ||
| + | |||
| + | ===THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP=== | ||
| + | |||
| + | {{ABSTRACT_PUBMED_1349852}} | ||
| + | |||
| + | ==About this Structure== | ||
| + | [[2pol]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. The June 2012 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Sliding Clamps'' by David Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2012_6 10.2210/rcsb_pdb/mom_2012_6]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2POL OCA]. | ||
| + | |||
| + | ==See Also== | ||
| + | *[[DNA polymerase|DNA polymerase]] | ||
| + | |||
| + | ==Reference== | ||
| + | <ref group="xtra">PMID:001349852</ref><ref group="xtra">PMID:016955075</ref><references group="xtra"/> | ||
| + | [[Category: DNA-directed DNA polymerase]] | ||
| + | [[Category: Escherichia coli]] | ||
| + | [[Category: RCSB PDB Molecule of the Month]] | ||
| + | [[Category: Sliding Clamps]] | ||
| + | [[Category: Kong, X P.]] | ||
| + | [[Category: Kuriyan, J.]] | ||
| + | [[Category: Nucleotidyltransferase]] | ||
| + | |||
== Introduction == | == Introduction == | ||
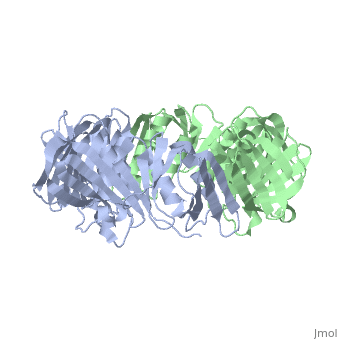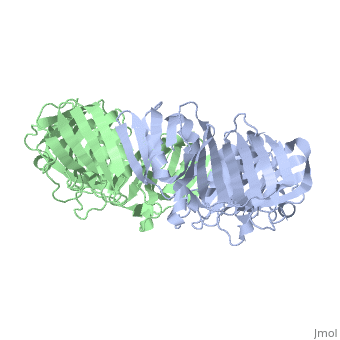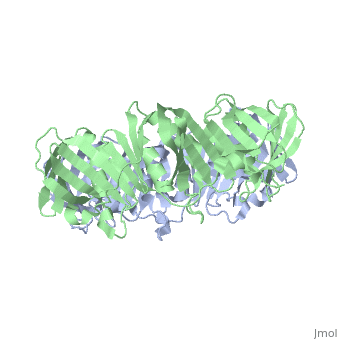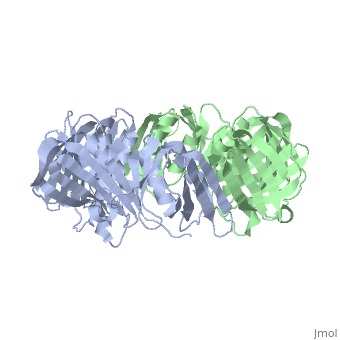
The <scene name='Polymerase_III_homoenzyme_beta_subunit/Beta_subunit_structure/1'>beta subunit</scene> of the Polymerase III holoenzyme is one of ten subunits in the Pol III multisubunit complex, which catalyzes the synthesis of both the leading and lagging strands of DNA in E. Coli. While the core Pol III enzyme (consisting of the alpha, sigma and theta subunits) dissociates from the DNA after replicating approximately 12 residues, the holoenzyme complex has a processivity of more than 5,000 residues. The increase in replication processing is due to the beta subunit. | The <scene name='Polymerase_III_homoenzyme_beta_subunit/Beta_subunit_structure/1'>beta subunit</scene> of the Polymerase III holoenzyme is one of ten subunits in the Pol III multisubunit complex, which catalyzes the synthesis of both the leading and lagging strands of DNA in E. Coli. While the core Pol III enzyme (consisting of the alpha, sigma and theta subunits) dissociates from the DNA after replicating approximately 12 residues, the holoenzyme complex has a processivity of more than 5,000 residues. The increase in replication processing is due to the beta subunit. | ||
Revision as of 09:41, 8 November 2012
Contents |
Polymerase III Holoenzyme beta subunit
THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP
Template:ABSTRACT PUBMED 1349852
About this Structure
2pol is a 2 chain structure with sequence from Escherichia coli. The June 2012 RCSB PDB Molecule of the Month feature on Sliding Clamps by David Goodsell is 10.2210/rcsb_pdb/mom_2012_6. Full crystallographic information is available from OCA.
See Also
Reference
- Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425-37. PMID:1349852
- Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006 Oct;7(10):751-61. Epub 2006 Sep 6. PMID:16955075 doi:10.1038/nrm2022
Introduction
The of the Polymerase III holoenzyme is one of ten subunits in the Pol III multisubunit complex, which catalyzes the synthesis of both the leading and lagging strands of DNA in E. Coli. While the core Pol III enzyme (consisting of the alpha, sigma and theta subunits) dissociates from the DNA after replicating approximately 12 residues, the holoenzyme complex has a processivity of more than 5,000 residues. The increase in replication processing is due to the beta subunit.
| |||||||||||