We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Josie N. Harmon/Sandbox Tutorial
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Xanthine Oxidoreductase == | == Xanthine Oxidoreductase == | ||
| - | Xanthine oxidoreductase is | + | Xanthine oxidoreductase is actually considered to exist as two enzymes in one, one being xanthine oxidase and the other existing as xanthine dehydrogenase. When the enzyme was initially purified by scientists two different forms of the enzyme were found, where one uses nicotinamide adenine dinucleotide (NAD) and the other uses oxygen. The two forms were believed to exist as two different enzymes and were thus given two different names; however, upon further investigation into the amino acid sequence of the enzymes it was determined that the enzymes were actually the same. The two forms of the enzyme can differ in two ways: |
1. Within the oxidoreductase enzyme there are multiple disulfide bridges. In the case that these bridges are left untouched the enzyme will then take on the characteristics to act as an oxidase, but in the event that these bridges are restricted the enzyme then acts as a dehydrogenase. | 1. Within the oxidoreductase enzyme there are multiple disulfide bridges. In the case that these bridges are left untouched the enzyme will then take on the characteristics to act as an oxidase, but in the event that these bridges are restricted the enzyme then acts as a dehydrogenase. | ||
| Line 11: | Line 11: | ||
== Mechanism of Action == | == Mechanism of Action == | ||
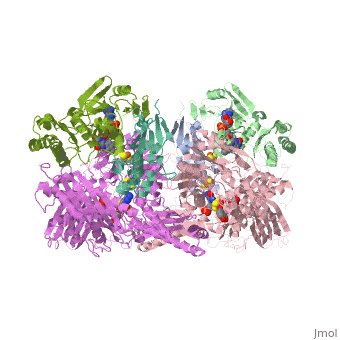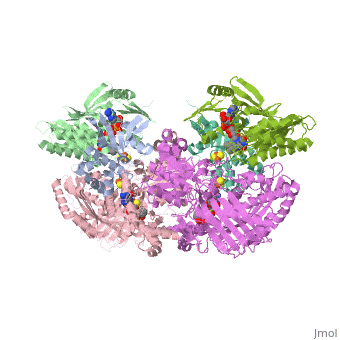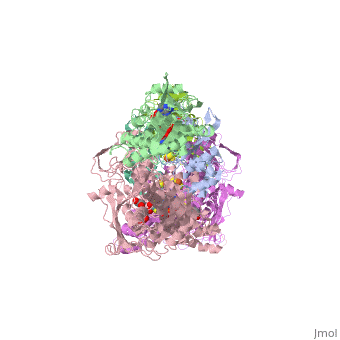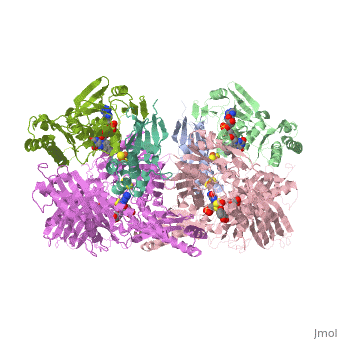
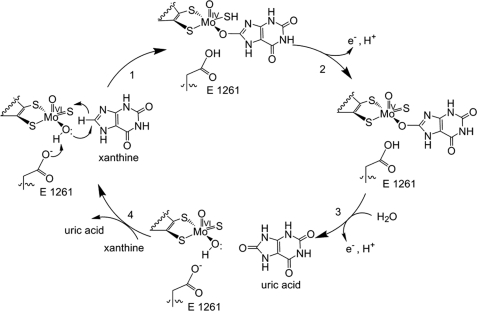
| - | Xanthine oxidase is characterized as a molybdenum containing enzyme that catalyzes the hydroxylation of a sp2 hybrized carbon in a broad range of aromatic heterocycles and aldehydes. In eukaryotes xanthine oxidase exist as a homodimer with each monomer containing four redox-active sites. The crystal structure of the bovine xanthine oxidase complex contains two active sites with varying intrinsic activity. The crystalline structure of a xanthine oxidase monomer offers a better view of the active molybdenum center, the ferredoxin iron sulfur, <scene name='User:Josie_N._Harmon/Sandbox_1/Fe2s2_name/1'>Fe2S2</scene>, clusters, and <scene name='User:Josie_N._Harmon/Sandbox_1/Fad_name/1'>FAD</scene>. The <scene name='User:Josie_N._Harmon/Sandbox_1/Active_site/1'>active site</scene> is thought to be composed of glutamine, glutamic acid, phenylalanine, arginine, and the molybdenum center. The substrate is believed to bind between the Phe 1009 and Phe 914. | + | Xanthine oxidase is characterized as a molybdenum containing enzyme that catalyzes the hydroxylation of a sp2 hybrized carbon in a broad range of aromatic heterocycles and aldehydes. In eukaryotes xanthine oxidase exist as a <scene name='User:Josie_N._Harmon/Sandbox_1/Xanthine_oxidoreductase/3'>homodimer</scene> with each monomer containing <scene name='User:Josie_N._Harmon/Sandbox_1/Ligands_monomer/4'>four redox-active sites</scene>. The crystal structure of the bovine xanthine oxidase complex contains two active sites with varying intrinsic activity. The crystalline structure of a xanthine oxidase monomer offers a better view of the active molybdenum center, the ferredoxin iron sulfur, <scene name='User:Josie_N._Harmon/Sandbox_1/Fe2s2_name/1'>Fe2S2</scene>, clusters, and <scene name='User:Josie_N._Harmon/Sandbox_1/Fad_name/1'>FAD</scene>. The <scene name='User:Josie_N._Harmon/Sandbox_1/Active_site/1'>active site</scene> is thought to be composed of glutamine, glutamic acid, phenylalanine, arginine, and the molybdenum center. The substrate is believed to bind between the Phe 1009 and Phe 914. |
== Electron Extraction == | == Electron Extraction == | ||
Revision as of 19:26, 12 November 2012
Xanthine Oxidase Biochemistry Tutorial
The purpose of this tutorial is to explain the mechanism of the metabolic enzyme xanthine oxidoreductase.
| |||||||||||