RA Mediated T-reg Differentiation
From Proteopedia
| Line 3: | Line 3: | ||
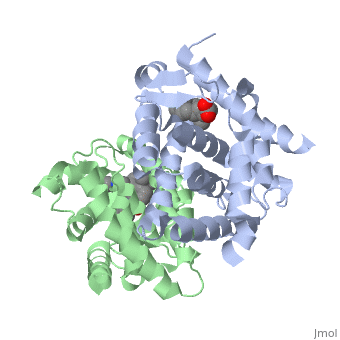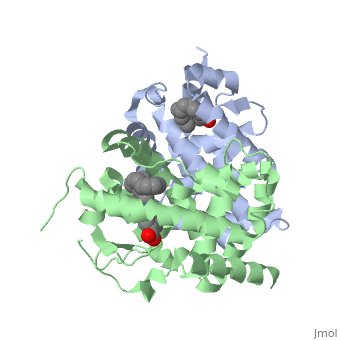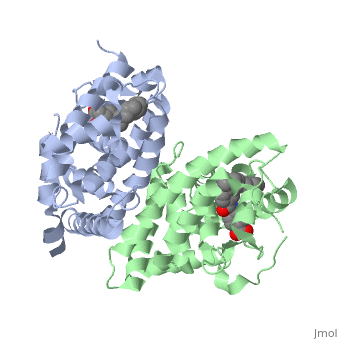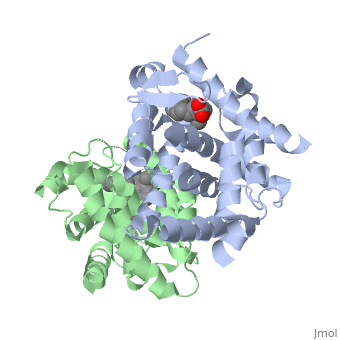
<StructureSection load='1dkf' size='350' side='left' caption='RARα-RXRα interaction (PDB entry [[1dkf]])' scene=''> | <StructureSection load='1dkf' size='350' side='left' caption='RARα-RXRα interaction (PDB entry [[1dkf]])' scene=''> | ||
==Ligand Binding Domain== | ==Ligand Binding Domain== | ||
| - | The Ligand binding domain for each piece of the dimer has a nearly identical structure of an <scene name='RA_Mediated_T-reg_Differentiaition/Alpha-helical_domains/2'>Tα-helical sandwich</scene>. These alpha helices form a total of 12 domains per protein (referred to as H1-12), with an additional 2 beta sheets as well. Additionally, the α-helical sandwich formed has been shown to bind All-Trans Retinoic Acid (ATRA), the isomer of RA used by the body. Both monomers contain two regions of activity, the <scene name='RA_Mediated_T-reg_Differentiaition/Dimerization_interface/3'>dimerization interface</scene> and the <scene name='RA_Mediated_T-reg_Differentiaition/Ligand_binding_pockets/1'> ligand binding pocket </scene> .<ref> PMID: 10882070 </ref> | + | The Ligand binding domain for each piece of the dimer has a nearly identical structure of an <scene name='RA_Mediated_T-reg_Differentiaition/Alpha-helical_domains/2'>Tα-helical sandwich</scene>. These alpha helices form a total of 12 domains per protein (referred to as H1-12), with an additional 2 beta sheets as well. Additionally, the α-helical sandwich formed has been shown to bind All-Trans Retinoic Acid (ATRA), the isomer of RA used by the body. Both monomers contain two regions of activity, the <scene name='RA_Mediated_T-reg_Differentiaition/Dimerization_interface/3'>dimerization interface</scene> and the <scene name='RA_Mediated_T-reg_Differentiaition/Ligand_binding_pockets/1'> ligand binding pocket </scene>.<ref> PMID: 10882070 </ref> |
| Line 30: | Line 30: | ||
Residue 270: α:Ile β:Ile γ:Met; Residue 232: α:Ser β:Ala γ:Ala; Residue 395: α:Val β:Val γ:Ala | Residue 270: α:Ile β:Ile γ:Met; Residue 232: α:Ser β:Ala γ:Ala; Residue 395: α:Val β:Val γ:Ala | ||
| - | The <scene name='RA_Mediated_T-reg_Differentiaition/Rxr-ligand_binding_pocket/1'>RXR-alpha binding pocket</scene> is comprised of 16 primarily hydrophobic residues. The ligand used in the crystal, Oleic Acid, is similar to RA, and RA is capable of binding to the RXRα pocket. | + | The <scene name='RA_Mediated_T-reg_Differentiaition/Rxr-ligand_binding_pocket/1'>RXR-alpha binding pocket</scene> is comprised of 16 primarily hydrophobic residues. The ligand used in the crystal, Oleic Acid, is similar to RA, and RA is capable of binding to the RXRα pocket.<ref> PMID: 10882070 </ref> |
Revision as of 08:11, 14 November 2012
Introduction
T-regulatory cells (T-regs) are a small subset of CD4+ T-cells that exhibit strong down regulation of immune system activity in their local environment. They are distinguished from other CD4+ T-cells by the expression of FOXP3, a gene regulator. [1] The exact mechanisms used by T-regs to down regulate the immune system has not yet been clearly elucidated. These cells have been shown to differentiate from CD4+ T-helper cells upon activation and exposure to the following cytokines: tumor growth factor β (TGF-β), Interleukin-2 (IL-2) and retinoic acid (RA). [2] Both TGF-β and IL-2 are used in other immune system differentiation, however, RA has been shown to bias T-cells to the T-reg phenotype. [3] When acting upon T-reg cells, RA acts as the ligand for the Retinoic Acid Receptor-α (RARα) / Retinoid X Receptor-α (RXRα) heterodimer. This heterodimer is of the nuclear receptor family, and each chain consists of the same three part structure: a Ligand binding domain (LBD), a DNA binding domain (DBD), and a hinge region connecting the two binding domains. [4]
| |||||||||||
| |||||||||||
Biological Significance
References
- ↑ Ochs HD, Oukka M, Torgerson TR. TH17 cells and regulatory T cells in primary immunodeficiency diseases. J Allergy Clin Immunol. 2009 May;123(5):977-83; quiz 984-5. PMID:19410687 doi:10.1016/j.jaci.2009.03.030
- ↑ Moore C, Fuentes C, Sauma D, Morales J, Bono MR, Rosemblatt M, Fierro JA. Retinoic acid generates regulatory T cells in experimental transplantation. Transplant Proc. 2011 Jul-Aug;43(6):2334-7. PMID:21839265 doi:10.1016/j.transproceed.2011.06.057
- ↑ Moore C, Fuentes C, Sauma D, Morales J, Bono MR, Rosemblatt M, Fierro JA. Retinoic acid generates regulatory T cells in experimental transplantation. Transplant Proc. 2011 Jul-Aug;43(6):2334-7. PMID:21839265 doi:10.1016/j.transproceed.2011.06.057
- ↑ Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999 May;64(5):310-9. PMID:10406480
- ↑ Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000 Feb;5(2):289-98. PMID:10882070
- ↑ Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000 Feb;5(2):289-98. PMID:10882070
- ↑ Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000 Feb;5(2):289-98. PMID:10882070
Proteopedia Page Contributors and Editors (what is this?)
William Bailey, Alexander Berchansky, Michal Harel, Jaime Prilusky