Sandbox Reserved 647
From Proteopedia
| Line 26: | Line 26: | ||
In relation to the eukaryotic dehydrogenase complex, bacteria have a smaller version of the complex where there are only 24 (E2) enzymes that make up the core. The core of the complex is arranged in a cubical shape with 8 vertices with each vertex being occupied by an (E2) trimer. The (E2) subunit of the complex is evolutionarily related to that of the eukaryotic enzyme. However the (E1) subunit found in bacteria is unrelated to that of the eukaryotic (E1) subunit. Bacterial (E1) is homodimer with a molecular weight of 99,474 Daltons containing <scene name='Sandbox_Reserved_647/Secondary_structure/1'> α/β folds </scene>and falls in the class of alpha and beta proteins. | In relation to the eukaryotic dehydrogenase complex, bacteria have a smaller version of the complex where there are only 24 (E2) enzymes that make up the core. The core of the complex is arranged in a cubical shape with 8 vertices with each vertex being occupied by an (E2) trimer. The (E2) subunit of the complex is evolutionarily related to that of the eukaryotic enzyme. However the (E1) subunit found in bacteria is unrelated to that of the eukaryotic (E1) subunit. Bacterial (E1) is homodimer with a molecular weight of 99,474 Daltons containing <scene name='Sandbox_Reserved_647/Secondary_structure/1'> α/β folds </scene>and falls in the class of alpha and beta proteins. | ||
== Function and Mechanism == | == Function and Mechanism == | ||
| - | [[Image:Mechanism photo.jpg | | + | [[Image:Mechanism photo.jpg | 350 px]] |
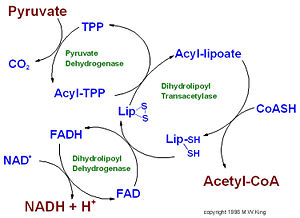
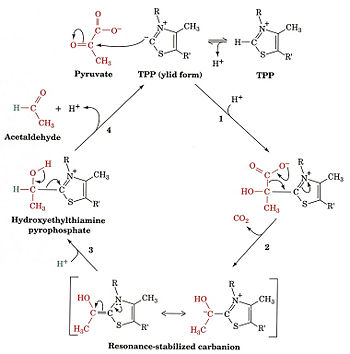
this is the site of <scene name='Sandbox_Reserved_647/Residue_interaction_with_mg/2'>interaction</scene> between three residues of pyruvate dehydrogenase. | this is the site of <scene name='Sandbox_Reserved_647/Residue_interaction_with_mg/2'>interaction</scene> between three residues of pyruvate dehydrogenase. | ||
Revision as of 00:36, 15 November 2012
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | |||||||||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
IntroductionPyruvate dehydrogenase (E1) along with two other enzymes make up the pyruvate dehydrogenase multienzyme complex. Dihydrolipoyl transacetylase (E2) and dihydrolipoyl dehydrogenase (E3) are the other two main components of the complex. Along with the associated prosthetic groups, Thiamine pyrophosphate (TPP), Lipoamide, and Flavin Adenine Dinucleotide (FAD) respective to each enzyme. These enzymes work together to synthesize acetyl Co-A by the irreversible oxidative decarboxylation of pyruvate before entering the Kreb's cycle. This catalytic reaction also produces NADH and carbon dioxide as side products. The pyruvate dehydrogenase complex acts to link glycolysis to the citric acid cycle. The pyruvate dehydrogenase complex is localized in the matrix space of the mitochondria in eukaryotes and the cytosol of bacteria. This complex is responsible for regulating the flow of energy in cells by determining when pyruvate should be converted into acetyl Co-A ore neutralized to lactic acid to allow continued glycolysis.
StructureEukaryotes The pyruvate dehydrogenase complex is the largest multienzyme complex known to exist with a molecular weight of about 9 Mega Daltons. The core of this multienzyme complex is composed of 60 (E2) subunits that form the shape of a pentagonal dodecahedron. This shape of composed of 20 vertices, with each vertex being occupied by an (E2) trimer. Each of these subunits has a linker region containing the lipoamide swinging arm that has contact with the (E1) subunits that surround the inner core. The outer shell of the complex is composed of 60 (E1) subunits. Each enzyme contacts one of the underlying (E2) enzymes. The (E3) enzyme lies in the center of the pentagon formed by the core of the (E2) enzymes and are associated with small binding proteins that are part of the complex. The pyruvate dehydrogenase enzyme itself is a tetramer that consists of two alpha subunits and two beta subunits and is present in 30 copies. Pyruvate dehydrogenase consists of a combination of beta strands and alpha helices. Prokaryotes In relation to the eukaryotic dehydrogenase complex, bacteria have a smaller version of the complex where there are only 24 (E2) enzymes that make up the core. The core of the complex is arranged in a cubical shape with 8 vertices with each vertex being occupied by an (E2) trimer. The (E2) subunit of the complex is evolutionarily related to that of the eukaryotic enzyme. However the (E1) subunit found in bacteria is unrelated to that of the eukaryotic (E1) subunit. Bacterial (E1) is homodimer with a molecular weight of 99,474 Daltons containing and falls in the class of alpha and beta proteins. Function and Mechanism
RegulationThe pyruvate dehydrogenase complex is regulated through reversible phosphorylation. The phosphorylation events occur on three serine residues at sites that are located on the pyruvate dehydrogenase (E1) alpha subunit. Once all three of the serine residues are phosphorylated the entire complex is inhibited, thus preventing the synthesis of Acetyl-CoA. The phosphorylation events are catalyzed by the enzyme pyruvate dehydrogenase kinase (PDK). Pyruvate dehydrogenase kinase exists in four know isoforms PDK1-PDK4. These kinases are distributed differently in various tissues. Activation of these kinases are also regulated by different factors. PDK1 is activated when there is a low concentration of oxygen available. PDK2 is activated when there is a high amount of Acetyl-CoA and NADH already present. PDK3 is activated when there is a high abundance of ATP already accessible for use. PDK4 is activated when the cell is suffering from starvation and nutrient deprivation. Dephosphorylation of the serine residues to restore the activity of the pyruvate dehydrogenase complex is catalyzed by the enzyme pyruvate dehydrogenase phosphatase (PDP). This enzyme has two known isoforms PDP1 and PDP2. PDP1 is prevalent in skeletal muscles and PDP2 is prevalent in the liver and adipocytes. |