Sandbox Reserved 390
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='3qx3' size='600' side='right' caption='Structure of the human topoisomeraseIIbcore-DNA cleavage complex stabilized by the anticancer drug etoposide. (PDB entry 3QX3)' scene=''> | <StructureSection load='3qx3' size='600' side='right' caption='Structure of the human topoisomeraseIIbcore-DNA cleavage complex stabilized by the anticancer drug etoposide. (PDB entry 3QX3)' scene=''> | ||
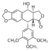
The purpose of this page is to explain a semisynthetic derivative of podophyllotoxin [[Image:PODO.png|thumb|right|50 px|Chemical structure of podophyllotoxin]] (etoposide) that demonstrates antitumor activity by inhibiting DNA topoisomerase II, thereby inhibiting DNA re-ligation which lead to apoptosis of the cancer cell and also, to describe the mechanisms of resistance of etoposide. | The purpose of this page is to explain a semisynthetic derivative of podophyllotoxin [[Image:PODO.png|thumb|right|50 px|Chemical structure of podophyllotoxin]] (etoposide) that demonstrates antitumor activity by inhibiting DNA topoisomerase II, thereby inhibiting DNA re-ligation which lead to apoptosis of the cancer cell and also, to describe the mechanisms of resistance of etoposide. | ||
| + | |||
__TOC__ | __TOC__ | ||
| - | |||
| - | |||
==INTRODUCTION == | ==INTRODUCTION == | ||
The research team described the structural basis by which an anticancer drug etoposide kills cancer cells by interacting with its cellular targets human DNA topoisomerase type II<ref>PMID: 21778401</ref>. In the close-up representation of the <scene name='Sandbox_Reserved_390/Top/21'>etoposide-binding site(s),</scene> cartoon-and-stick representation shows the insertion of two etoposide molecules into two cleavage sites [etoposide surrounded by orange mesh that represent active site (etoposide in red & grey representation), the DNA chain is in red and blue, and the magnesium is in green]. | The research team described the structural basis by which an anticancer drug etoposide kills cancer cells by interacting with its cellular targets human DNA topoisomerase type II<ref>PMID: 21778401</ref>. In the close-up representation of the <scene name='Sandbox_Reserved_390/Top/21'>etoposide-binding site(s),</scene> cartoon-and-stick representation shows the insertion of two etoposide molecules into two cleavage sites [etoposide surrounded by orange mesh that represent active site (etoposide in red & grey representation), the DNA chain is in red and blue, and the magnesium is in green]. | ||
Revision as of 22:15, 19 November 2012
Human topoisomerase IIbeta in complex with DNA and etoposide
| |||||||||||
References
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID:21778401 doi:10.1126/science.1204117
- ↑ Kathryn L. Gilroy, Chrysoula Leontiou, Kay Padget, Jeremy H. Lakey and Caroline A. Austin* "mAMSA resistant human topoisomerase IIβ mutation G465D has reduced ATP hydrolysis activity” Oxford JournalsLife Sciences Nucleic Acids Research Volume 34, Issue 5Pp. 1597-1607. DOI: 10.1093/nar/gkl057
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID:21778401 doi:10.1126/science.1204117