User:Sruti Aiyaswamy/Sandbox 1
From Proteopedia
| Line 29: | Line 29: | ||
==MR1-restricted MAIT Activation== | ==MR1-restricted MAIT Activation== | ||
<Structure load='4DZB' size='250' frame='true' align='right' caption='MAIT Cell Receptor' scene='' /> MAIT cells compose up to 10% of the peripheral blood T-cell population in humans. They are mostly associated with mesenteric lymph nodes and the gastrointestinal mucosa.<ref>PMID:12634786</ref> They interact with the MR1-antigen complex via their αβ T-cell antigen receptor (TCR). These TCRs consist of an α and β chain, and each chain has a constant (C) and variable (V) domains.<ref>PMID: 22412157</ref> The V domain of each chain has three complimentarity determining regions (CDRs). Together, the six CDRs form a ligand-binding site for the MAIT TCR. The MAIT TCR is restricted to MR1-antigen presentation. Mutational data suggests that a limited number of Vα chain residues of the MAIT TCR are critical for MR1-induced activation.<ref>PMID: 22412157</ref> | <Structure load='4DZB' size='250' frame='true' align='right' caption='MAIT Cell Receptor' scene='' /> MAIT cells compose up to 10% of the peripheral blood T-cell population in humans. They are mostly associated with mesenteric lymph nodes and the gastrointestinal mucosa.<ref>PMID:12634786</ref> They interact with the MR1-antigen complex via their αβ T-cell antigen receptor (TCR). These TCRs consist of an α and β chain, and each chain has a constant (C) and variable (V) domains.<ref>PMID: 22412157</ref> The V domain of each chain has three complimentarity determining regions (CDRs). Together, the six CDRs form a ligand-binding site for the MAIT TCR. The MAIT TCR is restricted to MR1-antigen presentation. Mutational data suggests that a limited number of Vα chain residues of the MAIT TCR are critical for MR1-induced activation.<ref>PMID: 22412157</ref> | ||
| - | <br> | + | <br> |
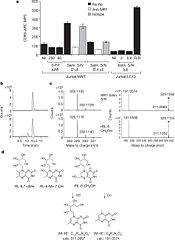
| - | + | Even though 6-FP is a ligand for MR1, it does not activate MAIT cells. It was found that antigen-presenting cells transfected with MR1 and infected with ''Salmonella typhimurium'' specifically activated MAIT cells. Notably, the supernatent from ''S. typhimurium'' was able to activate MAIT cells. Activation was assayed by CD69 upregulation and intracellular cytokine staining for interferon (IFN)-γ and tumor necrosis factor (TFN).<ref>PMID:23051753</ref> By searching for ligands from the supernatent that complexed with MR1, it was revealed that candidate MAIT-activating ligands are derivatives of riboflavin (vitamin B2).<ref>PMID:23051753</ref> These vitamin metabolites are structurally close to 6-FP, but they possess an extra ribityl moiety that may permit direct contact by the MAIT TCR. These compounds are derived from the riboflavin biosynthetic pathway present in most, but not all, bacteria and yeast. | |
[[Image:MAIT-cell antigens.jpg|175px|left|thumb|Identification of bacterially-derived MAIT-cell antigens]] | [[Image:MAIT-cell antigens.jpg|175px|left|thumb|Identification of bacterially-derived MAIT-cell antigens]] | ||
Revision as of 18:11, 27 November 2012
Contents |
Introduction
The MHC class I-like molecule, MR1, is an antigen presenting protein that specifically activates mucosal-associated invariant T (MAIT) cells. It has been found that microbial vitamin B metabolites serve as MR1 ligands that activate the MAIT cells.
MR1 is MHC Class I Related
|
Finally relate how MHC structure/function is similar to MR1. Explain chains, b2m, folding.
Structure of MR1-antigen Complex
| |||||||||||
MR1-restricted MAIT Activation
|
Even though 6-FP is a ligand for MR1, it does not activate MAIT cells. It was found that antigen-presenting cells transfected with MR1 and infected with Salmonella typhimurium specifically activated MAIT cells. Notably, the supernatent from S. typhimurium was able to activate MAIT cells. Activation was assayed by CD69 upregulation and intracellular cytokine staining for interferon (IFN)-γ and tumor necrosis factor (TFN).[4] By searching for ligands from the supernatent that complexed with MR1, it was revealed that candidate MAIT-activating ligands are derivatives of riboflavin (vitamin B2).[5] These vitamin metabolites are structurally close to 6-FP, but they possess an extra ribityl moiety that may permit direct contact by the MAIT TCR. These compounds are derived from the riboflavin biosynthetic pathway present in most, but not all, bacteria and yeast.
Biological Significance
There are a wide range of microbes that stimulate MAIT cells via MR1 interaction, and they all synthesize riboflavin. This suggests a possible mode by which MAIT cells might sense microbial infection or overgrowth at mucosal sites in an MR1-restricted manner.[6] There are possibly many species of bacteria in the gut flora that have riboflavin biosynthetic pathways. Perhaps the relatively frequent stimulation of MAIT cells serves as way for the body to strengthen the immune system, so as to not be overcome by the co-existing microbiota. The human microbiome is very delicately balanced. It is possible that a spiked growth of an opportunistic pathogen could trigger extensive MAIT cell activation (via MR1) to cause an effective immune response against the pathogen. There also could be other microbial-specific metabolites[7]that may serve as indicators of microbial infections that undergo immunosurveillance.
The pterin ring, an important component of MR1 ligands, occurs widely in nature and also represents a common scaffold of small molecule therapeutics.[8] Further research must be done to determine if MAIT cell-MR1 interactions are affected by drugs and diet.
References
- ↑ Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003 Mar 13;422(6928):164-9. PMID:12634786 doi:10.1038/nature01433
- ↑ Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012 Mar 12. PMID:22412157 doi:10.1084/jem.20112095
- ↑ Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012 Mar 12. PMID:22412157 doi:10.1084/jem.20112095
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605
- ↑ Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012 Jun 8;336(6086):1262-7. Epub 2012 Jun 6. PMID:22674330 doi:10.1126/science.1223813
- ↑ Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012 Oct 10. doi: 10.1038/nature11605. PMID:23051753 doi:10.1038/nature11605