Tutorial:Basic Chemistry Topics
From Proteopedia
| Line 1: | Line 1: | ||
| - | <div style='background-color:yellow;padding:20px;margin:20px;'><big>'''This tutorial is designed for entry-level college students with some basic chemistry knowledge (Ages 18-22)</big></div> | + | <StructureSection <div style='background-color:yellow;padding:20px;margin:20px;'><big>'''This tutorial is designed for entry-level college students with some basic chemistry knowledge (Ages 18-22)</big></div> |
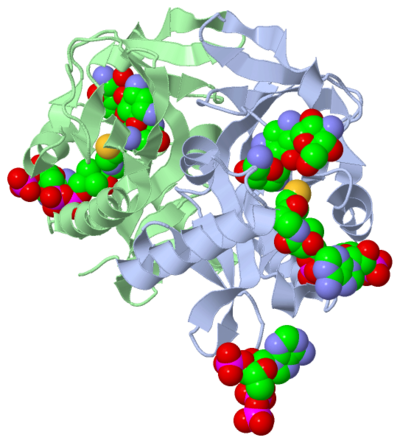
[[Image:1m4d.png|left|400px]]<ref name="Main Image">Vetting, M. W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin." RCSB Protien DataBase. N.p., 28 Aug.2002. Web. 13 July 2011. http://www.rcsb.org/pdb/explore/explore.do?structureId=1M4D</ref> | [[Image:1m4d.png|left|400px]]<ref name="Main Image">Vetting, M. W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin." RCSB Protien DataBase. N.p., 28 Aug.2002. Web. 13 July 2011. http://www.rcsb.org/pdb/explore/explore.do?structureId=1M4D</ref> | ||
| Line 19: | Line 19: | ||
| - | + | load='1m4d' size='600' side> | |
=='''Objectives'''== | =='''Objectives'''== | ||
Revision as of 23:44, 30 November 2012
This tutorial is designed for entry-level college students with some basic chemistry knowledge (Ages 18-22)</div> [1]
Purpose of the Tutorial
Summary: Scientific Research ArticleThe molecule to left is from the article "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin" published in Nature Structural Biology.[2]. The study focused on aminoglycoside 2’- N- acetyltransferase (AAC (2’)- Ic), an enzyme. This enzyme is a protein that speeds the rate of the reaction it catalyzes. This study determined the structure of AAC (2’)-Ic from Mycobacterium tuberculosis, a pathogen. This pathogen is a microorganism that causes tuberculosis (TB), which typically affects the lungs, but can affect other parts of the body as well. The specific structure/protein fold of AAC (2’)-Ic places it in the GCN5-related N-acetyltransferase (GNAT) superfamily. The GNAT superfamily is a group of enzymes that are similar in structure. The protein fold is important because it determines the function of the compound.[2] The GNAT family is a group of acetylating enzymes. Acetylation is the addition of CH3CO functional group onto a compound. Although the physiological function of AAC(2’)-Ic is not certain, the discovery of the GNAT fold allowed researchers to classify AAC (2’)-Ic as an acetylating enzyme. Mycothiol is catalyzed by AAC (2’)-Ic to acetylate the aminoglycoside antibiotic, Tobramycin. When this occurs the aminoglycoside antibiotic becomes inactive. The basis of this study is important because when pathogens become resistant or inactive to commonly used antibiotics, an infection that used to be easily cured can now become severe and life threatening.[2]
load='1m4d' size='600' side> ObjectivesBy the end of this tutorial you should be able to: 1. Describe and provide examples of covalent bonds, ionic bonds, and hydrogen bonds 2. Describe Primary Structure 3. Understand and identify the importance of secondary structures 4. Understand and describe an active site 4. Describe what a ligand does. Competitive/Allosteric inhibitors and inducers 7. Be able to classify amino acids and understand what the classification represents Periodic table
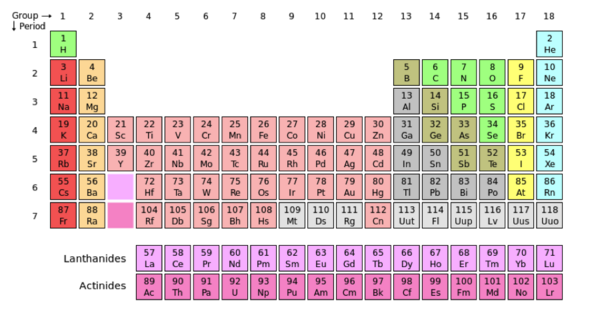
The image to the left displays Oxygen (O) in its periodic box. The atomic mass of oxygen is 8, centered above the single-letter abbreviation of Oxygen. “8” represents the number of protons in the atom’s nucleolus. The atomic number is also used to arrange elements in the periodic table, notice the increasing value of protons in the atoms from left to right. The atomic mass of an element is centered under the single-letter abbreviation of the atom and represents the total weight of the atom, in this case oxygen has an atomic mass of ~16. The total weight of an atom is the sum of the atoms protons, neutrons and electrons. The periodic table is a collaboration of atoms that are arranged according to their common features. Groups are the vertical columns of atoms, and the periods are the horizontal columns. When moving from left to right across the periods (horizontally), there is an increase in electronegativity. Electronegativity refers to the atoms ability to pull other electrons towards it, increasing its density and providing it’s self with a negative charge. The concept of electronegativity contributes to the polarity of a molecule. Polarity is caused by a difference in electronegativity between atoms in a compound and/or the asymmetry of the compounds structure. A compound is nonpolar when the electronegativity of the atoms/functional groups are close or even. The compound is symmetrical and the electrons are not being pulled more in one direction.
Amino acidsAmino acids are the building blocks of proteins. There are about 500 amino acids but we will be referencing the 20 more common ones. Polymer chains (peptides) join amino acids together to form proteins. The process of protein formation is known as translation. During translation, a complex known as the ribosome is responsible for adding amino acids to a peptide chain to form a complete protein. 20 More Common Animo Acids
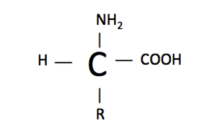
The basic structure of an amino acid is an amine group (-NH2), a carboxylic acid (-COOH), hydrogen (H), and a functional group (R). The functional group is what varies between amino acids. The functional group determines how the amino acid is categorized. They are categorized as essential/nonessential, polar/non-polar, and acidic/basic.
Classification of Amino AcidsEssentail vs. Nonessentail Amino Aicds An amino acid is considered essential when the human body is not capable of synthesizing/producing it. These amino acids need to be obtained from our diet. If the body is capable of producing an amino acid through metabolism or another method, the amino acid is classified as nonessential. Essential Amino Acids
Nonessential Amino Acids
The polarity of an amino acid is dependent on the difference in electronegativity and the asymmetry of the compounds structure, discussed previously. For example, Arginine is a polar amino acid and Glycine is a nonpolar amino acid. Neutral amino acids have functional groups that are similar in electronegativity, so the electrons are not pulled in one direction more dominantly then another. In other words, there is no increase in density to one side. When an amino acid is neutral, it is less reactive then a polar amino acid. It is less reactive because the structure is stable. A polar amino acid is pulling electrons yielding a slight positive and negative charge with in the amino acid structure, making it less stable. Those charges want to react with other atoms, yielding the higher reactivity. Nonpolar amino acids
Polar Amino Acids
Acidic vs. Basic Amino Acids
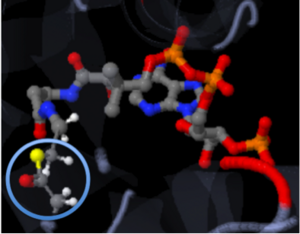
Types of BondsThere are three common types of bonds. These bonds include hydrogen bonds, covalent bonds, and ionic bonds. The strongest bond is a covalent bond, followed by the ionic bond, which leaves the weakest bond to be the hydrogen bond. Knowing where these bonds are utilized will aid in your understanding of why a structure is in a certain conformation. Covalent Bonds[4]The strongest type of bond is the covalent bond. Covalent bonds involve the sharing of electrons between two molecules/atoms. These bonds are very stable and are not easily broken. This represents Coenzyme A (CoA) with the addition of an acyl group to the sulfur. The acyl group is bound to CoA through covalent bonds. In the picture below, the acyl group is circled. All of the solid connecting bonds are covalent bonds.
Ionic Bonds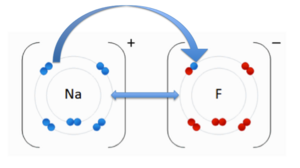 Ionic Bonding[5] An ionic bond is an attraction between two molecules of opposite charge. The opposite charge is either positive (+) or negative (-). A positively charged atom is referred to as a cation, and a negatively charged atom is referred to as an anion. In the image to the right, you see an anion, Fluorine (F) and the cation, Sodium (Na). These two atoms are attracted to each other due to their opposite charges. The double-sided arrow between them is representation of their attractive force. This representation highlights an ionic interaction between Tobramycin and Aspartic acid (Asp). The nitrogen on Tobramycin has a (+) positive charge and Aspartic acid has a (-) negative charge. The opposing charges are attracted to each other forming an ionic bond.
Hydrogen Bonds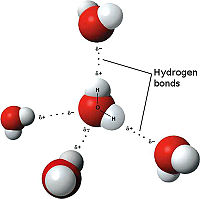 Hydrogen Bonding[6] The weakest bond, the hydrogen bond is an attractive interaction between an electronegative atom and hydrogen. Electronegative atoms have high electron density. High electron density refers to strong atoms that pull electrons towards it from weaker/low electron density atoms, such as hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. A common example of this is water. The image to the right demonstrates the hydrogen bonding in water. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons. When oxygen pulls the electrons, it leaves hydrogen with a slight positive charge. Since oxygen is pulling the hydrogen’s inward, the formation of a water droplet is possible. In this representation the hydrogen bonds are represented as yellow-dashed lines. The hydrogen bonds are important in this study and this molecular compound because they offer the stability of the secondary structures.
Primary StructureThe primary sequence of a compound is the amino acid linear sequence of the compound. For example the linear sequence of tobramycin is displayed below. The chemical formula is C18H37N5O9. If you count each of the elements the number should match. Image:Primary Structure.png Hydrogen Bonding
Secondary StructuresSecondary structures are alpha helices and beta sheets. The helices and sheets provide stability to the molecule as a whole. The alpha helices are represented with pink arrows and the beta strands are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. Alpha helices have a cylinder-like structure that rotates in a clockwise manner with a parallel formation. This representation shows these key features of alpha helices. Here, you can see the helices rotating clockwise (follow the arrows), and the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. Beta sheets are often anti-parallel, which are clearly represented in this figure. The folding of a protein, alpha helices and beta sheets, is what gives the compound its function. When there is a change in protein folding, the function will change.
Active SiteThe active site of a molecule can be described as a pocket where an interaction between compounds occurs. This interaction will cause a change in structure shape. The conformational change, change in structure shape, can inhibit or activate the physiological/pathological affect. The active site can either be inhibited or activated by the compound that binds there. Referring back to our article, the active site is where the acetylation is going to occur. In this depiction of the active site you can see the pocket where Coenzyme A (CoA) will aid the enzyme in the acetylation of Tobramycin (Toy), the aminoglycoside antibiotic. The enzyme is the "blob" surrounding the antibiotic and CoA. The enzyme/protein is holding these compounds in a conformation forcing them to react. The acetylation at the active site will cause the antibiotic to be inactive, hence inhibiting the active site. When Tobramycin becomes inactivated it is no longer able to aid in the destruction of bacteria. This is what we call antibiotic resistance.
SubstratesA substrate is a compound that is acted upon by the enzyme. The substrate binds the active site of the enzyme, once bound the enzyme transfers a functional group(s) to the substrate. After the substrate has collected the functional group(s) it is released from the active site. When this occurs the substrate can go on to bind the enzymes compound of interest and transfer the acquired functional groups to produce the product. Once the final transformation occurs the product is either inhibited or activated by the conformational change.[3]
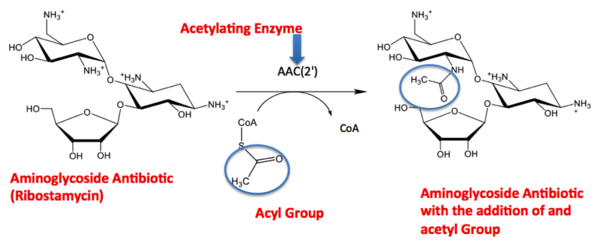 Acetylation Reaction [2]
LigandLigands are molecules or complexes that are located within the secondary structures. The ligands are held in a certain conformation by the secondary structure. This conformation contributes to the function of the compound, as mentioned before. Ligands can have binding sites on receptors. When bound to the receptor it can trigger a physiological response. A ligand can be a competitive agonist, allosteric agonist, competitive antagonist, or an allosteric antagonist. An agonist is a ligand that causes a physiological response, activating the active site. An antagonist is a ligand that inhibits a physiological response, not allowing the active site to be activated. A ligand is competitive when it is binding to the same site as the physiological activator; hence it is competing for the same site. When a ligand binds to an allosteric site, the ligand is binding to the same receptor but it is not binding to the active site. The ligands present in the research article complex are coenzyme A, Tobramycin and Phosphate-Adenosine-5'-Diphosphate. The ligands in this complex are represented as a dimer. A dimer is a chemical structure formed from two identical subunits. Some molecules are present as a dimer because it is more stable then the monomer, one subunit. The dimer is constructed by connecting two subunits along their axis.
The hide-aways boxes located below provide more information about CoA and Tobramycin for those who are interested.
http://www.elmhurst.edu/~chm/vchembook/561aminostructure.html
| ||||||||||||||||||
References
- ↑ Vetting, M. W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin." RCSB Protien DataBase. N.p., 28 Aug.2002. Web. 13 July 2011. http://www.rcsb.org/pdb/explore/explore.do?structureId=1M4D
- ↑ 2.0 2.1 2.2 2.3 2.4 Vetting, Matthew W., et al. "Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin."Nature Structural Biology 9.9 (2002): 653-58. Print.
- ↑ 3.0 3.1 3.2 Wikipedia. Wikipedia, 4 Nov. 2012. Web. 7 Nov. 2012. <http://en.wikipedia.org/wiki/Enzyme_substrate_(biology)
- ↑ User:Cepheus. "Periodic Table." Wikipedia. N.p., 26 Feb. 2007. Web. 26 Nov. 2012. <http://en.wikipedia.org/wiki/File:Periodic_table.svg>.
- ↑ . "File:NaF.gif." Wikipedia. Wikipedia, 17 June 2011. Web. 31 Oct. 2012.<http://en.wikipedia.org/wiki/File:NaF.gif.
- ↑ Maňas, Michal, trans. "File:3D model hydrogen bonds in water.jpg." Wikimedia Commons. Wikimedia Commons, 3 Dec. 2007. Web. 31 Oct. 2012 <http://commons.wikimedia.org/wiki/File:3D_model_hydrogen_bonds_in_water.jpg.
- ↑ "Tobramycin." Wikipedia. Wikipedia, n.d. Web. 26 Nov. 2012.<http://en.wikipedia.org/wiki/Tobramycin>.