Objectives
By the end of this tutorial you should be able to:
1. Describe and provide examples of covalent bonds, ionic bonds, and hydrogen bonds
2. Describe Primary Structure
3. Understand and identify the importance of secondary structures
4. Understand and describe an active site
4. Describe what a ligand does. Competitive/Allosteric inhibitors and inducers
7. Be able to classify amino acids and understand what the classification represents
The "element color code" drop down box maybe needed throughout the tutorial for identification of atoms.
| Element Color Code
|
|
Carbon- Grey
Nitrogen- Blue
Sulfur- Yellow
Oxygen- Red
|
Periodic table
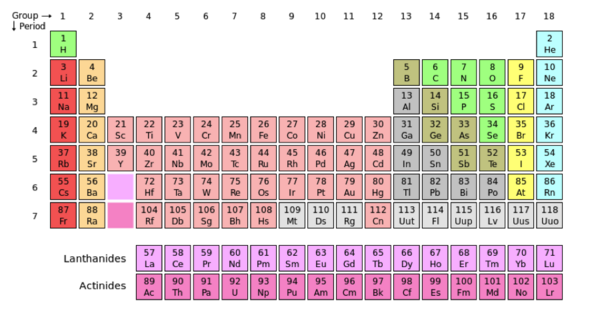
An atom is the smallest unit of matter. Protons, neutron, and electrons, make up the composition of an atom providing its physical features. The common physical features of atoms are used to organize them in the periodic table. Electrons are negatively charged particles that surround the atom’s nucleolus. Protons are positively charged particles and neutrons are uncharged. Both neutrons and protons are located in the central part of the atom known as the nucleus.
The image above displays Oxygen (O) in its periodic box. The atomic number of oxygen is 8, centered above the single-letter abbreviation of Oxygen. “8” represents the number of protons in the atom’s nucleus. The atomic number is also used to arrange elements in the periodic table. Notice the increasing value of protons in the atoms from left to right. The atomic mass of an element is centered under the single-letter abbreviation of an atom and represents the total weight of the atom. In this case, oxygen has an atomic mass of ~16. The total weight of an atom is the sum of the atoms protons, neutrons and electrons.
The periodic table is a collaboration of atoms that are arranged according to their common features. Groups are the vertical columns of atoms, and the periods are the horizontal rows. When moving from left to right across the periods (horizontally), there is an increase in electronegativity. Electronegativity refers to the atoms ability to pull other electrons towards it, increasing it’s density and providing itself with a negative charge. The concept of electronegativity contributes to the polarity of an atom. Polarity is caused by a difference in electronegativity between atoms in a compound and/or the asymmetry of the compounds structure. A compound is nonpolar when the electronegativity of the atoms/functional groups are close or even. The compound is symmetrical and the electrons are not being pulled more in one direction.
Amino acids
Amino acids are the building blocks of proteins. There are about 500 amino acids, but we will be referencing the 20 more common ones. Polymer chains (peptides) join amino acids together to form proteins. The process of protein formation is known as translation. During translation, a complex known as the ribosome is responsible for adding amino acids to a peptide chain to form a complete protein.
20 More Common Animo Acids
- Alanine (ala)(A)
- Glycine (gly)(G)
- Isoleucine (ile)(I)
- Leucine (leu)(L)
- Methionine (met) (M)
- Phenylalanine (phe)(F)
- Proline (pro)(P)
- Valine (val) (V)
- Arginine (arg)(R)
- Asparagine (asn)
- Aspartic acid (asp)(D)
- Cysteine (cys)
- Glutamic Acid (glu)(E)
- Glutamine (gln) (Q)
- Histidine (his)(H)
- Lysine (lys) (K)
- Serine (ser) (S)
- Threonine (thr)(T)
- Tryptophan (trp) (W)
- Tyrosine (tyr)(Y)
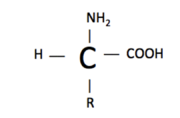
An amino acid structure consists of an amine group (-NH2), a carboxylic acid (-COOH), hydrogen (H), and a functional group (R). The functional group is what varies between amino acids and determines how it is categorized. Amino acids are categorized as either essential/nonessential, polar/non-polar, or acidic/basic.
Classification of Amino Acids
Essential vs. Nonessentail Amino Aicds
An amino acid is considered essential when the human body is not capable of synthesizing/producing it. These amino acids need to be obtained from our diet. If the body is capable of producing an amino acid through metabolism or another method, the amino acid is classified as nonessential.
Essential Amino Acids
- Histidine (his)
- Isoleucine (ile)
- Leucine (leu)
- Lysine (lys)
- Methionine (met)
- Phenylalanine (phe)
- Threonine (thr)
- Tryptophan (trp)
- Valine (val)
Nonessential Amino Acids
- Alanine(ala)
- Arginine (arg)
- Asparagine (asn)
- Aspartic acid, (asp)
- Cysteine (cys)
- Glutamic acid (glu)
- Glutamine (gln)
- Proline (pro)
- Serine (ser)
- Tyrosine (tyr)
Neutral, Polar vs. Nonpolar Amino Acids
The polarity of an amino acid is dependent on the difference in electronegativity and the asymmetry of the compounds structure, discussed previously. For example, Arginine is a polar amino acid and Glycine is a nonpolar amino acid. As we learned earlier, an amino acid’s structure consists of a carboxylic acid, an amine, hydrogen, and a functional group. Look at the structures of . Arginine has the amine group, carboxylic acid and hydrogen located towards the bottom of the representation, and the functional group is the large extension of atoms upward. You can see from this image that the functional group has a greater density/electronegativity compared to the core of the amino acid (carboxylic acid, amine and hydrogen), hence making this amino acid polar. In contrast the structure of glycine, located next to arginine, has little polarity. The functional group attached to glycine is only a methyl group (CH3). A methyl group has low density/electronegativity compared to the rest of the structure, making glycine a nonpolar amino acid. Neutral amino acids have functional groups that are similar in electronegativity compared to the core, so the electrons are not pulled in one direction more dominantly then another. In other words, there is no increase in density to one side. When an amino acid is neutral, it is less reactive then a polar amino acid. It is less reactive because the structure is stable. A polar amino acid is pulling electrons yielding a slight positive and negative charge within the amino acid structure, making it less stable. Those charges want to react with other atoms, yielding the higher reactivity.
Nonpolar amino acids
- Alanine (ala)
- Glycine (gly)
- Isoleucine (ile)
- Leucine (leu)
- Methionine (met)
- Phenylalanine (phe)
- Proline (pro)
- Valine (val)
Polar Amino Acids
- Arginine (arg)
- Asparagine (asn)
- Aspartic acid (asp)
- Cysteine (cyc)
- Glutamic Acid (glu)
- Glutamine (gln)
- Histidine (his)
- Lysine (lys)
- Serine (ser)
- Threonine (thr)
- Tryptophan (trp)
- Tyrosine (tyr)
Neutral Amino Acids
- Alanine (ala)
- Asparagine (asn)
- Cysteine (cyc)
- Glutamine (gln)
- Glycine (gly)
- Isoleucine (ile)
- Leucine (leu)
- Methionine (met)
- Phenylalanine (phe)
- Proline (pro)
- Serine (ser)
- Threonine (thr)
- Tryptophan (trp)
- Tyrosine (tyr)
- Valine (val)
Acidic vs. Basic Amino Acids
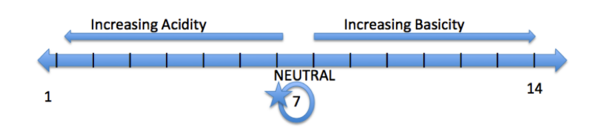
An Example of a pH scale is located above. A pH scale ranges from 1-14, 1 being the most acidic and 14 being the most basic. An acidic pH has a value below 7, a basic pH is greater than 7, and a pH of 7 is considered neutral (ex: water). Some common acidic functional groups are alcohols (OH) and carboxylic acids (COOH). Some basic functional groups are amines (NH3) and ethers (CH2OCH2).
You can predict if an amino acid will be acidic or basic according to its structure. For example, contains a carboxylic acid functional group, which was stated previously to be an acidic functional group that lowers the pH of the amino acid. On the other hand, is classified as a basic amino acid. The functional group of arginine contains a guanidine group. A guanidine group contains amine groups, increasing the pH of the amino acid.
Acidic Amino Acids
- Aspartic acid (asp)
- Glutamic acid (glu)
Basic Amino Acids
- Arginine (arg)
- Histidine (his)
- Lysine (lys)
Types of Bonds
There are many different types of bonds that occur physiologically. Ionic, covalent, and hydrogen bonds are some of the most abundantly seen. The strongest of these is the covalent bond, followed by the ionic bond, which leaves the weakest bond to be the hydrogen bond. Knowing where these bonds are utilized will aid in your understanding of why a structure is in a certain conformation.
Covalent Bonds
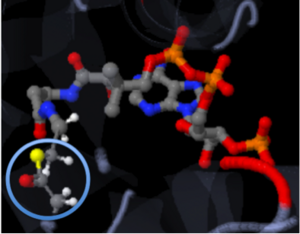
The strongest of these bonds is the covalent bond. Covalent bonds involve sharing pairs of electrons between two atoms. These bonds are very stable. The figure shown below, represents Coenzyme A (CoA) with the addition of an acyl group to the sulfur atom. The introduction mentioned that the addition of the acetyl group is important to the study because when it is transferred to Tobramycin, the antibiotic becomes inactive. The solid connections between the atoms are representation of the covalent bonds.
[4]
Ionic Bonds
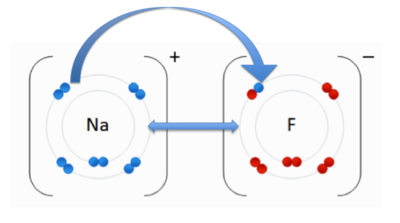
An ionic bond occurs when an electron(s) is transferred from one atom to another due to the opposite charges of the atoms. The positively charged cation is attracted to the negatively charged anion. In the image to the right, you see an anion, Fluorine (F) and the cation, Sodium (Na). The double-sided arrow between them is representation of their attractive force. Fluorine has a higher electronegativity than sodium. As we discussed previously, when an atom has higher electronegativity it pulls electrons from the lower electronegative atom, in this case sodium. The transfer of the sodium electron (blue) is shown using an arrow.
This representation highlights an between tobramycin and aspartic acid (Asp). The nitrogen on tobramycin has a (+) positive charge and aspartic acid has a (-) negative charge. The opposing charges are attracted to each other forming an ionic bond, holding the compounds in close proximity.
Hydrogen Bonds
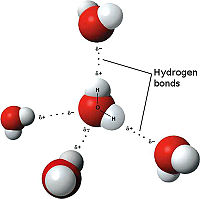
The weakest bond discussed here, the hydrogen bond is an attractive interaction between an electronegative atom and hydrogen. When the electronegative atom pulls the electrons, it leaves the other atom with a slightly positive charge. A common example of this is water. The image to the right demonstrates the hydrogen bonding of water. The highly electronegative oxygen pulls the hydrogen closer by attracting hydrogen’s electrons. When oxygen pulls the electrons, it leaves hydrogen with a slight positive charge. Since oxygen is pulling the hydrogen’s inward, the formation of a water droplet is possible. In this representation the are represented as yellow-dashed lines. The hydrogen bonds are important to the compound used by the study because they offer stability to the secondary structures.
Primary Structure
The primary sequence is the amino acid linear sequence of a protein. For example the linear sequence of the protein enzyme AAC (2’)-Ic is shown below.
The letters in the primary structure are the single-letter abbreviations of the amino acids. The single-letter abbreviations are provided in the amino acid portion of this tutorial. The primary sequence of AAC (2’)-Ic, a protein enzyme, was an important discovery to the study because it revealed the cause of the tobramycin acetylation, leading to its inactivity.
Secondary Structures
Secondary structures are . The helices and sheets provide stability to the compound as a whole. The alpha helices are represented with pink arrows and the beta sheets are represented with yellow arrows. This molecule has approximately eight alpha helices and four beta sheets. have a cylinder-like structure with a parallel formation. In this representation you can see the parallel formation within the cylinder structure. The parallel alpha helices are held in its cylinder structure by hydrogen bonds. are often anti-parallel. The folding of a protein, alpha helices and beta sheets, gives the compound it’s function. When there is a change in protein folding, the function will change. As previously stated, the study discovered ACC(2’)-Ic to have a GNAT fold, and the GNAT family are enzymes capable of acetylation.
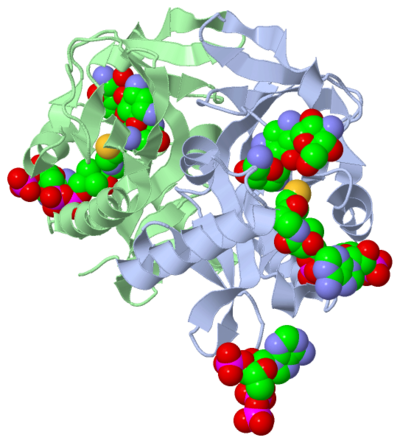
Active Site
The active site can be described as a pocket where an interaction between complexes produces a chemical reaction. The chemical reaction can be physiological or pathological. Referring back to our article, the active site is where the acetylation is going to occur. In this depiction of the you can see the pocket where coenzyme A (CoA) will aid the enzyme in the acetylation of tobramycin (Toy), the aminoglycoside antibiotic. The enzyme is the "blob" surrounding the antibiotic and CoA. The enzyme is holding these compounds in a conformation forcing them to react. The acetylation at the active site will cause the antibiotic to be inactive, hence inhibiting the active site. When tobramycin becomes inactivated it is no longer able to aid in the destruction of bacteria. This is what we call antibiotic resistance.
Substrates
A substrate is a compound that is acted upon by an enzyme. The substrate binds the active site of the enzyme. Once bound, the enzyme uses the substrate to produce a chemical response that produces the product. After the chemical response, the substrate is released from the active site and you are left with the enzyme and the product. [3]
This equation is a representation of the interaction between a substrate and an enzyme. The enzyme (E) and substrate (S) join together at the enzyme’s active site to form an enzyme-substrate (ES) complex. Once the enzyme releases the substrate you are left with the final product (P) and the enzyme (E).
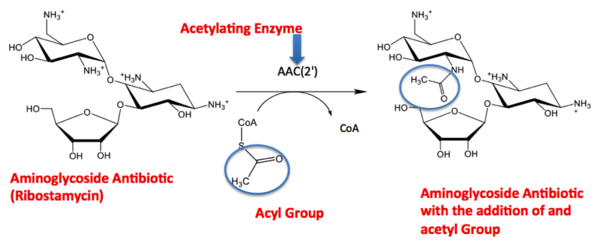
As discussed in the introduction, AAC(2’) Ic has a similar fold to that of the GNAT superfamily. The GNAT fold described in the study has the function of acetylation, the addition of an acetyl group. An acetyl functional group is composed of CH3CO. It is important to note that the discovery of the GNAT fold lead to the understanding of the function of AAC(2’). The reaction above shows the acetylation of the aminoglycoside antibiotic, causing its inactivity. From the reaction centered above you see the aminoglycoside antibiotic (Ribostamycin) being acted upon by the enzyme AAC(2’). AAC(2’) is adding and acetyl group to the antibiotic using the substrate CoA. On the right side of the arrow you can see the final product of the acetylation, the antibiotic and acyl group bound. The Acetyl group is circled, so you are able to locate it throughout the reaction. [2]
Ligand
are molecules or complexes located within the secondary structures. The ligands are held in a certain conformation by the secondary structure. This conformation contributes to the function of the compound, as mentioned earlier. The amino acids located throughout the protein will react with components of the ligands based on the polararity/nonpolarity and basicity/acidity of the ligand components. The ligands present in the research article complex are , and Phosphate-Adenosine-5'-Diphosphate.
The ligands in this complex are present as a . A dimer is a chemical structure formed with two identical subunits, connected along their axis. The dimer conformation is sometimes favored over the monomer due to an increase in stability.
The drop-down boxes below provide more information about the ligands present in the study article for those who are interested.
| Coenzyme A (CoA)
|
| Coenzyme (CoA) is a coenzyme that synthesizes and oxidizes fatty acids. This process is essential for the utilization of fatty acids. Coenzyme A is used as a substrate in the citric acid cycle. The citric acid cycle is also known as the Krebs cycle or tricarboxylic acid cycle (TCA). This process is important to the production of ATP, which is an energy source used by the body.
|
| Tobramycin
|
|
Tobramycin is an antibiotic that is part of the aminoglycoside family. Aminoglycosides produce antibacterial effects by inhibiting protein synthesis and compromising the cell wall structure. By inhibiting the protein synthesis of the bacteria, it does not allow the bacteria to replicate. The cell wall is an important structure to bacteria because it provides the structure and stability to the bacteria. By disrupting the cell wall, we are removing the stability of the bacteria and ultimately casing bacteria death. [7]
Tobramycin targets a variety of bacteria, particularly gram(-) species. Just like all drugs there are side effects associated with tobramycin. Some of the more common side effects are ototoxicity and nephrotoxicity. Ototoxicity is hearing loss and nephrotoxicity is causing kidney damage. The kidney damage is due to Tobramycin reabsorption through the renal tubules. This basically means that tobramycin may be toxic to the kidneys because of prolonged contact time in the kidneys.ref name="Tobramycin">
Tobramycin trade name is Tobrex. A trade name is another name for tobramycin. It is a pregnancy category D. Pregnancy categories are assigned to all drugs. They are used to classify how likely the drug is to cause harm to the fetus. The pregnancy categories are A, B, C, D, and X. Pregnancy category A causes no harm to the fetus and pregnancy category X indefinitely causes harm to the fetus. Since Tobramycin is a pregnancy category D, this is not an optimal choice for a pregnant patient.ref name="Tobramycin">
Tobramycin can be given intravenously, intramuscularly, as an inhalation or ophthalmicly. Intravenously is an IV route of administration where the drug is administered directly to the vasculature or blood vessels. Intramuscular is a shot that penetrates your muscle. A common example of an intramuscular administration would be the flu shot. Inhalation is a route of administration where the lungs are the targets. An example of this would be an inhaler used in asthmatics. Ophthalmic administration is where the drug is administered to the eye; an example would be an eye drop.ref name="Tobramycin">
|
Preventing Antibiotic Resistance
Drug resistance is when microorganisms are able to survive when in the presence of drugs that used to be effective in eliminating them. Resistance occurs when a microorganism produces a spontaneous/genetic mutation that protects the bacteria from the drug. Antibiotic resistance to Tobramycin by Mycobacterium tuberculosis is the basis of this study, as discussed earlier. Antibiotic resistance is a growing public health concern that needs to be addressed.
There are many causes of drug resistance, including antibiotic overuse and inappropriate antibiotic use. A preventable cause of drug resistance that the consumers should be aware of is prematurely discontinuing antibiotics. Patients tend to discontinue antibiotics when they start to “feel better” but the infection is not completely eradicated. When you begin an antibiotic regimen it begins to kill the most susceptible microorganisms first. As you continue the antibiotic the continuous exposure of the antibiotic is able to kill the few more resistant microorganisms. When the antibiotic is discontinued prior to the elimination of the more resistant microorganisms, those bacteria begin to replicate again and the same antibiotic will not be as effective.
When drug resistance occurs the drug/antibiotic needs to be modified to prevent its inactivation. In this study Tobramycin is acetylated, which causes it inactivation. If we can prevent the bacteria from acetylating Tobramycin, then Tobramycin can remain active and continue its antibacterial effect. Ways to inhibit an addition of an unwanted functional group or other additions, is to attach a bulky substituent or a compound with the same charge as the possible unwanted addition. Adding a bulky substance to Tobramycin will “block” the addition of the acetyl group, because the acetyl group cannot get to the site of action. If a compound of the same charge is added adjacent to where the unwanted addition is added. The two similar charges will be in close range to each other causing them to repel each other, like we discussed earlier in our discussion of ionic bonds.
In conclusion, understanding and applying the basic chemistry concepts discussed here can aid in future research that can benefit so many areas of study. You have the ability to change lives and provide the world with life changing discoveries.
http://www.elmhurst.edu/~chm/vchembook/561aminostructure.html