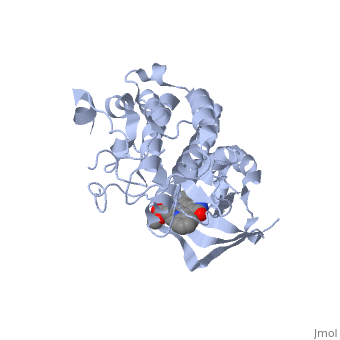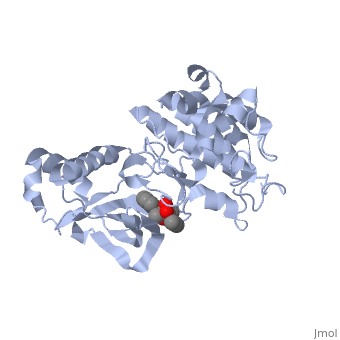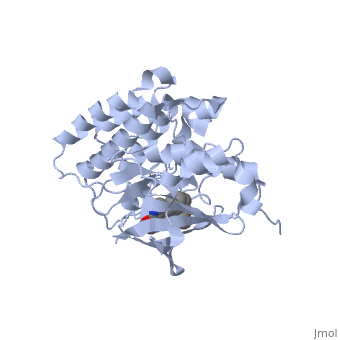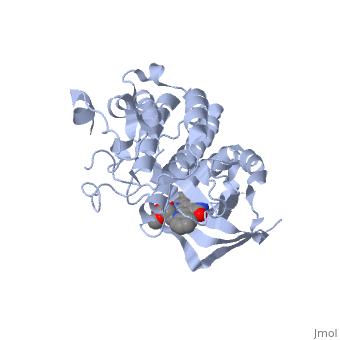
Hepatocyte growth factor receptor
From Proteopedia
| Line 29: | Line 29: | ||
<scene name='Hepatocyte_growth_factor_receptor/K-252a/1'>K-252a</scene> is a staurosporine analog. Staurosporine is an inhibitor of many Ser/Thr Kinases, and has been shown to also inhibit c-Met activation by inhibiting its autophosphorylation. The structures of K-252a and staurosporine are very similar, with the main difference being that K-252a has a furanose instead of a pyranose Moiety. The binding of K-252a causes the c-Met to adopt an inhibitory conformation of the A-loop, specifically with residues <scene name='Hepatocyte_growth_factor_receptor/Residues1231-1244/1'>1231-1244</scene> . This segment blocks the place where the substrate tyrosine side chain would bind, if the protein were in an active conformation. Residues | <scene name='Hepatocyte_growth_factor_receptor/K-252a/1'>K-252a</scene> is a staurosporine analog. Staurosporine is an inhibitor of many Ser/Thr Kinases, and has been shown to also inhibit c-Met activation by inhibiting its autophosphorylation. The structures of K-252a and staurosporine are very similar, with the main difference being that K-252a has a furanose instead of a pyranose Moiety. The binding of K-252a causes the c-Met to adopt an inhibitory conformation of the A-loop, specifically with residues <scene name='Hepatocyte_growth_factor_receptor/Residues1231-1244/1'>1231-1244</scene> . This segment blocks the place where the substrate tyrosine side chain would bind, if the protein were in an active conformation. Residues | ||
| - | <scene name='Hepatocyte_growth_factor_receptor/Catalytic_1127/1'>1223-1230</scene> also enhance this inhibitory conformation, as they | + | <scene name='Hepatocyte_growth_factor_receptor/Catalytic_1127/1'>1223-1230</scene> also enhance this inhibitory conformation, as they constrain C into a conformation that does not allow the catalytic placement of Glu-1127 keeping αC in an inactive conformation. |
| - | + | Residues 1229-1230 pass through the triphosphate subsite of bound ATP blocking ATP binding. The K-252a itself binds in the adenosine pocket, therefore also inhibiting the binding of ATP. The binding of K-252a is very favorable (enthalpy change of -17.9 kcal/mol). This is probably due to polar interactions as well as a change in conformation upon binding. <ref>PMID: 14559966</ref> | |
There is a concerteted conformational change in the complex upon K-252a binding. One of these changes involves the A-loop, specifically residues 1228-1230. In the Apo-Met structure, the side chain of Met-1229 would pass through the ring of the inhibitor, and so, in order to make room for K-252a, the segment must move, with residues 1229 and 1230 moving by 3-4 Å. In order to make room for the side chain of Tyr-1230, arg-1208 moves by 8 Å toward Asp-1204. Are-1208, which in the uninhibited complex would stack with tyr-1230, now stacks with phe-1234 <ref>PMID: 14559966</ref> | There is a concerteted conformational change in the complex upon K-252a binding. One of these changes involves the A-loop, specifically residues 1228-1230. In the Apo-Met structure, the side chain of Met-1229 would pass through the ring of the inhibitor, and so, in order to make room for K-252a, the segment must move, with residues 1229 and 1230 moving by 3-4 Å. In order to make room for the side chain of Tyr-1230, arg-1208 moves by 8 Å toward Asp-1204. Are-1208, which in the uninhibited complex would stack with tyr-1230, now stacks with phe-1234 <ref>PMID: 14559966</ref> | ||
| Line 43: | Line 43: | ||
C-Terminal Docking Site | C-Terminal Docking Site | ||
| - | There are | + | There are three binding motifs at the C-terminal supersite that act as docking sites in C-met. The first two are 1356YVNV and 1349YVHV, each (in the wild type protien) containing a tyrosine that gets phosphorylated. These tyrosines correspond to residues <scene name='Hepatocyte_growth_factor_receptor/Tyrisine_docking_sites/1'>1349 and 1356</scene>. In this structure, these residues are inhibited from becoming phosphorylated. The residues <scene name='Hepatocyte_growth_factor_receptor/Extended_conformation/1'>1349-1352</scene> form an extended conformation, which is seen in other phosphopeptides that bind to SH2 domians. Residues 1353-1356 form a type I β turn, which is similar to sequences that bind to Shc-PTB domians. Whether binding to SH2 domains or PTB domains, upon binding, these motifs would move to avoid clashes with the C lobe. The third binding motif is found in residues 1356-1359, which form a type II β turn, and is similar to pohsphopeptides that bind Grb2. These three binding motifs of the mutated structure are very similar to binding motifs that would be recognized by their binding parters, implying that the C-terminal supersite of this structure is very similar to that of an active c-met. <ref>PMID: 14559966</ref> |
<scene name='Hepatocyte_growth_factor_receptor/Beta_turn/1'>β turns</scene>. | <scene name='Hepatocyte_growth_factor_receptor/Beta_turn/1'>β turns</scene>. | ||
==Biological Significance== | ==Biological Significance== | ||
| - | Many human cancers can be traced back to mutations in the c-met kinase domain, and these mutations often lead to over activation of this kinase. | + | Many human cancers can be traced back to mutations in the c-met kinase domain, and these mutations often lead to over activation of this kinase. Some of the following mutations are known to cause an over active c-met: V1092I, H1094L/Y/R, H1106D, M1131T, V1188L, L1195V, V1220I, D1228H/N, Y1230H/C, Y1235D, K1244R, and M1250T/I. Many of these mutations most likely affect the A loop conformation of the wild type receptor, causing it to become constitutively active. This is done by either stabilizing the active form of the enzyme or destabilizing the inactive form. |
| + | |||
| + | The significance of this particular structure is that it shows the A loop of c-Met is flexible and will adapt in order to bind to an indolocarbazole (K-252a). This gives insight on designs for specific c-Met inhibitors, as this specific site can be targeted by drugs to block c-Met activity. | ||
| + | |||
| + | This structure also shows the binding motifs of c-Met in an unphosphorylated form, giving insight on how the motifs may move when interacting with their respective binding domains (Grb2, SH2, PTB domains). | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Revision as of 21:04, 4 December 2012
Contents |
Hepatocyte Growth Factor Receptor
| |||||||||||
Mutated Receptor in Complex with K-252a
|
Mutation
This particular structure of the hepatocyte growth factor tyrosine kinase domain is one harboring a human cancer mutation. The two are replaced by a phenylalanine and aspartate, respectively. This mutation normally causes the receptor to be consitutively active, and is found in metastatic HNSC carcinoma. Although there is no longer phosphorylation at these sites, it is believed that the aspartate negative charge resembles the negative phosphate that would normally cause activation, and therefore keeps the protein in its active form. [10] There is a third mutation at Tyr-1194 which is substituted for a . This is shown to point into the formed by Lys-1198 and Leu-1195 from αE. [11] This structure is conserved in the wild type protein, suggesting that the mutation at residue 1149 is not changing the structure at this position.
K-252a
is a staurosporine analog. Staurosporine is an inhibitor of many Ser/Thr Kinases, and has been shown to also inhibit c-Met activation by inhibiting its autophosphorylation. The structures of K-252a and staurosporine are very similar, with the main difference being that K-252a has a furanose instead of a pyranose Moiety. The binding of K-252a causes the c-Met to adopt an inhibitory conformation of the A-loop, specifically with residues . This segment blocks the place where the substrate tyrosine side chain would bind, if the protein were in an active conformation. Residues also enhance this inhibitory conformation, as they constrain C into a conformation that does not allow the catalytic placement of Glu-1127 keeping αC in an inactive conformation. Residues 1229-1230 pass through the triphosphate subsite of bound ATP blocking ATP binding. The K-252a itself binds in the adenosine pocket, therefore also inhibiting the binding of ATP. The binding of K-252a is very favorable (enthalpy change of -17.9 kcal/mol). This is probably due to polar interactions as well as a change in conformation upon binding. [12]
There is a concerteted conformational change in the complex upon K-252a binding. One of these changes involves the A-loop, specifically residues 1228-1230. In the Apo-Met structure, the side chain of Met-1229 would pass through the ring of the inhibitor, and so, in order to make room for K-252a, the segment must move, with residues 1229 and 1230 moving by 3-4 Å. In order to make room for the side chain of Tyr-1230, arg-1208 moves by 8 Å toward Asp-1204. Are-1208, which in the uninhibited complex would stack with tyr-1230, now stacks with phe-1234 [13]
K-252a binds in the adenosine pocket. It has four hydrogen bonds to the enzyme, with of these mimicking hydrogen bonds of an adenine base. There is a hydrogen bond between the lactam nitrogen and the carbonyl oxygen of Pro-1158, and another between the lactam carbonyl oxygen and the hydrogen of the amide of Met-1160. There are two more hydrogen bonds between the 3' hydroxyl and carbonyl oxygen and the of the A loop. [14]
There are also many hydrophobic interactions between the interface of the enzyme and K-252a. The residues involved in this are Ile-1084, Gly-1085, Phe-1089, Val-1092, Ala-1108, Lys-1110, and Leu-1140 (); Leu-1157, Pro-1158, Tyr-1159, and Met-1160 (); and Met-1211, Ala-1226, Asp-1228, Met-1229, and Tyr-1230 (). [15]
Met-1229, Met-1211 and Met-1160 all make up the for the indolocarbazole plane as they are all within van der waals distance of it. [16]
C-Terminal Docking Site
There are three binding motifs at the C-terminal supersite that act as docking sites in C-met. The first two are 1356YVNV and 1349YVHV, each (in the wild type protien) containing a tyrosine that gets phosphorylated. These tyrosines correspond to residues . In this structure, these residues are inhibited from becoming phosphorylated. The residues form an extended conformation, which is seen in other phosphopeptides that bind to SH2 domians. Residues 1353-1356 form a type I β turn, which is similar to sequences that bind to Shc-PTB domians. Whether binding to SH2 domains or PTB domains, upon binding, these motifs would move to avoid clashes with the C lobe. The third binding motif is found in residues 1356-1359, which form a type II β turn, and is similar to pohsphopeptides that bind Grb2. These three binding motifs of the mutated structure are very similar to binding motifs that would be recognized by their binding parters, implying that the C-terminal supersite of this structure is very similar to that of an active c-met. [17] .
Biological Significance
Many human cancers can be traced back to mutations in the c-met kinase domain, and these mutations often lead to over activation of this kinase. Some of the following mutations are known to cause an over active c-met: V1092I, H1094L/Y/R, H1106D, M1131T, V1188L, L1195V, V1220I, D1228H/N, Y1230H/C, Y1235D, K1244R, and M1250T/I. Many of these mutations most likely affect the A loop conformation of the wild type receptor, causing it to become constitutively active. This is done by either stabilizing the active form of the enzyme or destabilizing the inactive form.
The significance of this particular structure is that it shows the A loop of c-Met is flexible and will adapt in order to bind to an indolocarbazole (K-252a). This gives insight on designs for specific c-Met inhibitors, as this specific site can be targeted by drugs to block c-Met activity.
This structure also shows the binding motifs of c-Met in an unphosphorylated form, giving insight on how the motifs may move when interacting with their respective binding domains (Grb2, SH2, PTB domains).
References
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ http://en.wikipedia.org/wiki/C-Met
- ↑ Maina F, Casagranda F, Audero E, Simeone A, Comoglio PM, Klein R, Ponzetto C. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell. 1996 Nov 1;87(3):531-42. PMID:8898205
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100
- ↑ Schiering N, Knapp S, Marconi M, Flocco MM, Cui J, Perego R, Rusconi L, Cristiani C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12654-9. Epub 2003 Oct 14. PMID:14559966 doi:10.1073/pnas.1734128100