Molecular Playground/Nickel Superoxide Dismutase
From Proteopedia
| Line 19: | Line 19: | ||
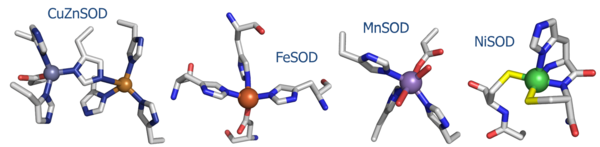
[[Image:SODs.png|600 px|thumb|'''Fig. 1:''' Active site structures of the four known SOD's.]] | [[Image:SODs.png|600 px|thumb|'''Fig. 1:''' Active site structures of the four known SOD's.]] | ||
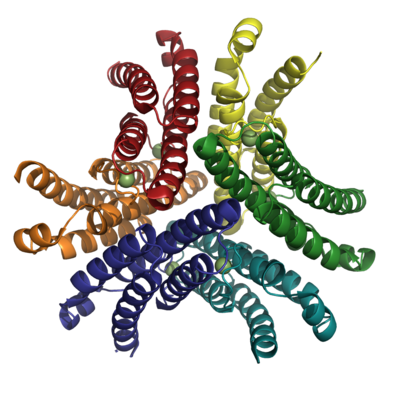
| - | [[Image:NiSOD Hexamer.png| | + | [[Image:NiSOD Hexamer.png|400 px|thumb|Fig. 2: Hexameric NiSOD. Figure created with PyMol. PDB ID 1T6U.]] |
Revision as of 21:22, 11 December 2012
Molecular Playground/Nickel Superoxide Dismutase
Nickel superoxide dismutase (NiSOD) is one of the CBI Molecules being studied in the University of Massachusetts, Amherst Chemistry-Biology Interface Program at Umass Amherst and on display at the Molecular Playground.
Introduction
Nickel Superoxide Dismutase (NiSOD) is the newest member in a class of enzymes that protects organisms from oxidative stress caused by superoxide, a harmful free radical byproduct of aerobic metabolism. NiSOD reacts with two molecules of superoxide, to form O2 and H2O2 with rates occurring at or near the diffusion limit. During catalysis, the redox-active nickel center cycles between an oxidized and reduced state. This reaction is termed the pin pong mechanism and is shown below.
- M(n + 1) + O2•- → Mn+ + O2
Mn+ + O2•- + 2H+ → M(n + 1) + H2O2
—————————————————
- 2O2•- + 2H+ → O2 + H2O2
NiSOD is distinct among SOD's for three reasons:
(1) NiSOD shares no sequence homology with the other known SOD's (Fig. 1).
(2) The ligands employed in the redox-active metal center are distinct. Cu/Zn, Fe, and MnSOD employ only aspartic acids, waters, and histidines. In NiSOD, the nickel center is coordinated by the side chains of cysteine 2 and cysteine 6, as well as the N-terminal amine, the amide group of cysteine 2 and an axial histidine ligand.
(3) Copper, iron, and manganese are redox active in aqueous media at biological pH. Nickel does not, and requires the coordination of the two cysteine ligands to tune its redox potential.