TAL effector
From Proteopedia
| Line 22: | Line 22: | ||
==DNA binding module with marked RVD== | ==DNA binding module with marked RVD== | ||
| - | <!--[[Image:3D_sanjana.png|frame|DNA binding module with marked RVD | ||
| - | <ref>[http://en.wikipedia.org/wiki/Image:3D_Influenza_virus.png Image of influenza virus structure] was obtained from Wikipedia.</ref>.]] | ||
| - | --> | ||
<br><br>[[Image:sanjana.png]]<br><br> | <br><br>[[Image:sanjana.png]]<br><br> | ||
'''Figure 1''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = T, HD = C, NI = A, and NN = G or A. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site. | '''Figure 1''': Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = T, HD = C, NI = A, and NN = G or A. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site. | ||
| + | |||
| + | ==Engineering== | ||
| + | <br>[[Image:iGEM.png]]<br> | ||
| + | '''Figure 3''': Based on the RVD the construction of a so called GATE Assembly kit was performed by the iGem 2012 Team from Freiburg University. | ||
==Reference== | ==Reference== | ||
Revision as of 16:05, 2 January 2013
Transcription activator-like effector PthXo1 (TALE PthXo1)TALEs are proteins originally derived from the plant pathogene Xanthomonas spp. The gene encoding for the effector are Pthxo1 and Pxo_00227. Originally they were detected in Xanthomonas Oryzae but the TALE PthXo1 genes were transformed into Escherichia Coli to express the DNA-binding protein.
Contents |
3D structures of TALE PthXo1
2jg2 – PpSPT + aldimine – Pseudomonas paucimobilis
2jgt – PpSPT
2w8j - SpSPT + aldimine – Sphingomonas paucimobilis
2w8t, 2w8u, 2w8v, 2w8w - SpSPT (mutant) + aldimine
2xbn – SpSPT + pyridoxamine phosphate
3a2b – SPT + pyridoxal phosphate + serine – Sphingobacterium multivorum
About this Structure
DNA binding module with marked RVD
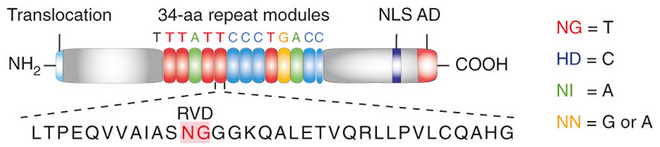
Figure 1: Natural structure of TALEs derived from Xanthomonas sp. Each DNA-binding module consists of 34 amino acids, where the RVDs in the 12th and 13th amino acid positions of each repeat specify the DNA base being targeted according to the cipher NG = T, HD = C, NI = A, and NN = G or A. The DNA-binding modules are flanked by nonrepetitive N and C termini, which carry the translocation, nuclear localization (NLS) and transcription activation (AD) domains. A cryptic signal within the N terminus specifies a thymine as the first base of the target site.
Engineering
Image:IGEM.png
Figure 3: Based on the RVD the construction of a so called GATE Assembly kit was performed by the iGem 2012 Team from Freiburg University.
Reference
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target. Science. 2012 Jan 5. PMID:22223736 doi:10.1126/science.1216211
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012 Jan 5;7(1):171-92. doi: 10.1038/nprot.2011.431. PMID:22222791 doi:10.1038/nprot.2011.431
Proteopedia Page Contributors and Editors (what is this?)
Philipp Warmer, Michal Harel, Jaime Prilusky, OCA, Alexander Berchansky
