We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2qsg
From Proteopedia
(Difference between revisions)
m (Protected "2qsg" [edit=sysop:move=sysop]) |
|||
| Line 1: | Line 1: | ||
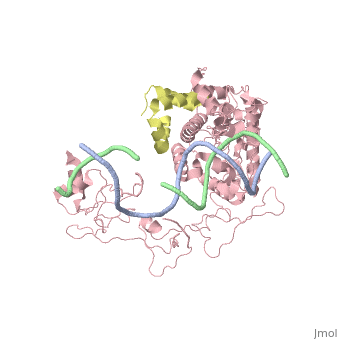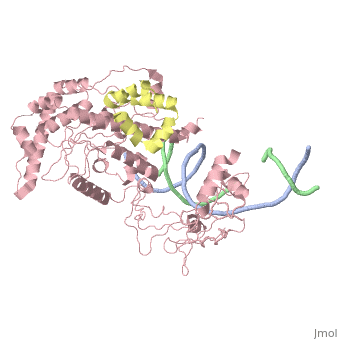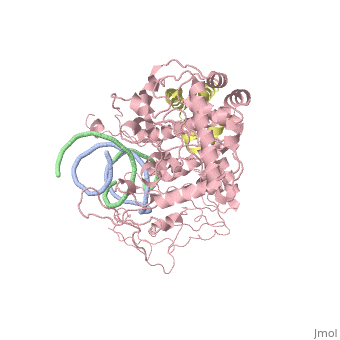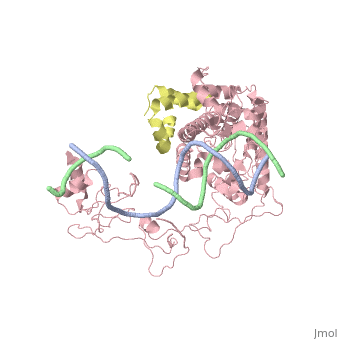
| - | [[ | + | ==Crystal structure of Rad4-Rad23 bound to a UV-damaged DNA== |
| + | <StructureSection load='2qsg' size='340' side='right' caption='[[2qsg]], [[Resolution|resolution]] 3.10Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[2qsg]] is a 4 chain structure with sequence from [http://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2QSG OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2QSG FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=N:ANY+5-MONOPHOSPHATE+NUCLEOTIDE'>N</scene></td></tr> | ||
| + | <tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[2qsf|2qsf]], [[2qsh|2qsh]]</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">RAD4 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae]), RAD23 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 Saccharomyces cerevisiae])</td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2qsg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2qsg OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2qsg RCSB], [http://www.ebi.ac.uk/pdbsum/2qsg PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/qs/2qsg_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Mutations in the nucleotide excision repair (NER) pathway can cause the xeroderma pigmentosum skin cancer predisposition syndrome. NER lesions are limited to one DNA strand, but otherwise they are chemically and structurally diverse, being caused by a wide variety of genotoxic chemicals and ultraviolet radiation. The xeroderma pigmentosum C (XPC) protein has a central role in initiating global-genome NER by recognizing the lesion and recruiting downstream factors. Here we present the crystal structure of the yeast XPC orthologue Rad4 bound to DNA containing a cyclobutane pyrimidine dimer (CPD) lesion. The structure shows that Rad4 inserts a beta-hairpin through the DNA duplex, causing the two damaged base pairs to flip out of the double helix. The expelled nucleotides of the undamaged strand are recognized by Rad4, whereas the two CPD-linked nucleotides become disordered. These findings indicate that the lesions recognized by Rad4/XPC thermodynamically destabilize the Watson-Crick double helix in a manner that facilitates the flipping-out of two base pairs. | ||
| - | + | Recognition of DNA damage by the Rad4 nucleotide excision repair protein.,Min JH, Pavletich NP Nature. 2007 Oct 4;449(7162):570-5. Epub 2007 Sep 19. PMID:17882165<ref>PMID:17882165</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | + | ||
| - | == | + | |
| - | < | + | |
[[Category: Saccharomyces cerevisiae]] | [[Category: Saccharomyces cerevisiae]] | ||
[[Category: Min, J H.]] | [[Category: Min, J H.]] | ||
Revision as of 19:02, 30 September 2014
Crystal structure of Rad4-Rad23 bound to a UV-damaged DNA
| |||||||||||
Categories: Saccharomyces cerevisiae | Min, J H. | Pavletich, N P. | Alpha-beta structure | Beta hairpin | Cyclobutanepyrimidine cpd dimer | Dna binding | Dna binding protein | Dna binding protein-dna complex | Dna repair | Dna-damage recognition | Mismatch dna | Nucleotide excision repair | Protein-dna complex | Transglutaminase fold | Ultraviolet uv damage | Xeroderma pigmentosum