Tropomyosin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1c1g' size='450' side='right' scene='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_dimer/2' caption=''> | |
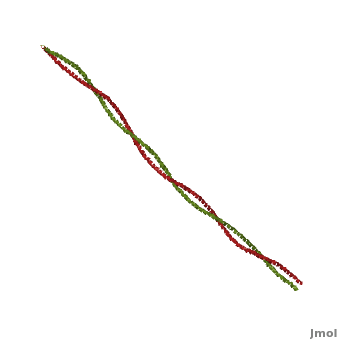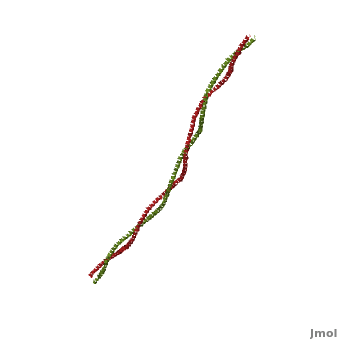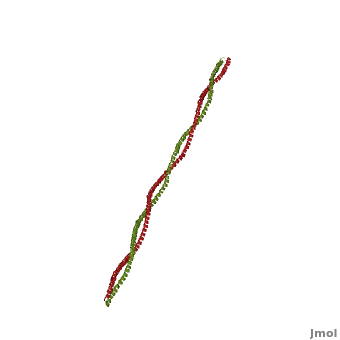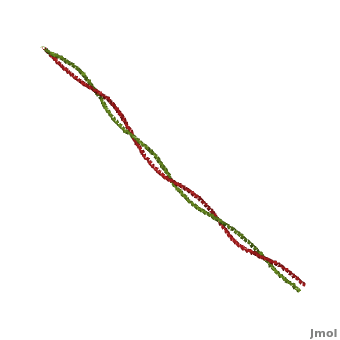
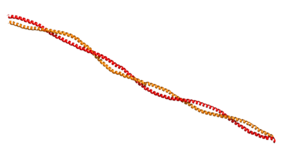
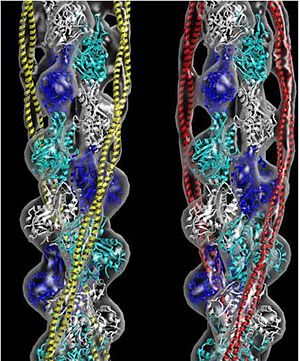
[[Image:Tropomyosin Dimer-2.png | thumb | left| 300x180px | alt text | '''Tropomyosin:''' Coiled-Coil Dimer, which is composed of two alpha helices [http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G (1C1G)] ]][[Image:B Lehman1.jpg | thumb | 300 x 430px | left | alt text | '''Tropomyosin''' (seen in yello and red) wrapped around actin filaments, which are EM reconstructions with G-actin ribbion structures filling in the EM structure. (Picture generated from William Lehman's [http://www.bumc.bu.edu/phys-biophys/research/filhel/ Website]) ]]'''[[Tropomyosin]] (TM)''' is an [[actin]] binding protein, which consists of a coiled-coil dimer (see left) and forms a polymer along the length of actin by a head-to-tail overlap along the major grove of actin (see down & left)<ref name="Gunning">Tropomyosins. I. Gunning, Peter, 1950- II. Series.[DNLM: 1. Tropomyosin. W1 AD559 v.644 2008 / WE 500 T856 2008]</ref>. The head-to-tail overlap allows flexibility between the tropomyosin dimers so it will lay unstrained along the filament<ref name="Gunning"/>. Each tropomyosin molecule spans seven actin monomers within a filament and lays N- to C- terminally from actin's pointed to barbed end<ref name="Frye">PMID:20465283</ref>. The 284 amino acid helix has a length of 420 Angstroms and has a molecular weight around 65-70 kilodaltons (vertebrate tropomyosin)<ref name="Gunning"/><ref name="Whitby">PMID:10651038</ref>. A few of tropomyosin's characteristics as an actin binding protein includes regulation, stabilization and recruitment. | [[Image:Tropomyosin Dimer-2.png | thumb | left| 300x180px | alt text | '''Tropomyosin:''' Coiled-Coil Dimer, which is composed of two alpha helices [http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G (1C1G)] ]][[Image:B Lehman1.jpg | thumb | 300 x 430px | left | alt text | '''Tropomyosin''' (seen in yello and red) wrapped around actin filaments, which are EM reconstructions with G-actin ribbion structures filling in the EM structure. (Picture generated from William Lehman's [http://www.bumc.bu.edu/phys-biophys/research/filhel/ Website]) ]]'''[[Tropomyosin]] (TM)''' is an [[actin]] binding protein, which consists of a coiled-coil dimer (see left) and forms a polymer along the length of actin by a head-to-tail overlap along the major grove of actin (see down & left)<ref name="Gunning">Tropomyosins. I. Gunning, Peter, 1950- II. Series.[DNLM: 1. Tropomyosin. W1 AD559 v.644 2008 / WE 500 T856 2008]</ref>. The head-to-tail overlap allows flexibility between the tropomyosin dimers so it will lay unstrained along the filament<ref name="Gunning"/>. Each tropomyosin molecule spans seven actin monomers within a filament and lays N- to C- terminally from actin's pointed to barbed end<ref name="Frye">PMID:20465283</ref>. The 284 amino acid helix has a length of 420 Angstroms and has a molecular weight around 65-70 kilodaltons (vertebrate tropomyosin)<ref name="Gunning"/><ref name="Whitby">PMID:10651038</ref>. A few of tropomyosin's characteristics as an actin binding protein includes regulation, stabilization and recruitment. | ||
| Line 14: | Line 14: | ||
#'''Superfamily:''' tropomyosin | #'''Superfamily:''' tropomyosin | ||
#'''Family:''' pig [[http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G 1c1g]] | #'''Family:''' pig [[http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G 1c1g]] | ||
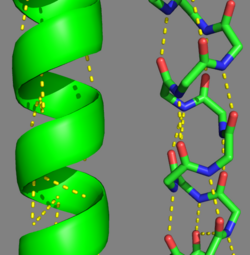
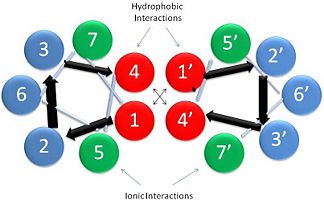
| - | + | [[Image:Helical Wheel Respresentation of Tropomyosin.jpg | thumb | right | 333x200px | alt text | '''Helical Wheel Diagram Representation of Tropopomyosin:''' The coiled-coil dimer is stabilized by hydrophobic and ionic interactions (red=hydrophobic, blue=polar & green=charged)Image Reconstructed from <ref name="Gunning"/>.]] Categorization of tropomyosin's <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_dimer/2'>coiled-coil</scene> describes a unique pattern of amino acids within the primary structure of the alpha helices that comprise the dimer interface<ref name="Gunning"/>. The unique amino acid pattern, found within all coiled-coil proteins, is a heptad repeat, which follows a similar pattern to: '''H-P-P-H-C-P-C''', where H is hydrophobic, P is polar and C is charged<ref name="Gunning"/><ref name="Whitby"/>. This heptad repeat forms a right-handed alpha helical secondary structure (see right for alpha helix secondary structure)<ref name="Gunning"/>. This alpha helix is special in that it forms a hydrophobic strip along one side, which will interface with an adjacent alpha helix that also contains the heptad repeat and hydrophobic strip. These strips aid in the dimerization of tropomyosin and is important in the characteristic coiled-coil domain. | |
<br /> | <br /> | ||
This coiled-coil domain is represented as a helical wheel diagram (see right), whereby the four amino acids (two from each alpha chain) that are adjacent to each other contribute to a <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_hydrophobic_aa/4'>hydrophobic interaction</scene>, represented in red, while the four amino acids (two from each alpha chain) flanking the hydrophobic core provide an ionic interaction, or salt-bridge represented as green. The <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_ionic_interaction/2'>salt-bridge</scene> can be observed by observing chain A, residue 26, which is a glutamic acid and forms a salt bridge with an arginine, 305 on Chain B. This weak stabilizing interaction has both and electrostatic interaction, via the ions, and two hydrogen bonds between the residues. This extra stability aids in its left-handed super-coil conformation. The two hydrogen bonds between the residues have an average distance of 2.85Å. Combined the hydrophobic and ionic interactions contribute significantly to the stability of the dimer<ref name="Gunning"/><ref name="Whitby"/>. | This coiled-coil domain is represented as a helical wheel diagram (see right), whereby the four amino acids (two from each alpha chain) that are adjacent to each other contribute to a <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_hydrophobic_aa/4'>hydrophobic interaction</scene>, represented in red, while the four amino acids (two from each alpha chain) flanking the hydrophobic core provide an ionic interaction, or salt-bridge represented as green. The <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_ionic_interaction/2'>salt-bridge</scene> can be observed by observing chain A, residue 26, which is a glutamic acid and forms a salt bridge with an arginine, 305 on Chain B. This weak stabilizing interaction has both and electrostatic interaction, via the ions, and two hydrogen bonds between the residues. This extra stability aids in its left-handed super-coil conformation. The two hydrogen bonds between the residues have an average distance of 2.85Å. Combined the hydrophobic and ionic interactions contribute significantly to the stability of the dimer<ref name="Gunning"/><ref name="Whitby"/>. | ||
Revision as of 12:44, 1 May 2013
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Gregory Hoeprich, Jaime Prilusky, David Canner, Joel L. Sussman