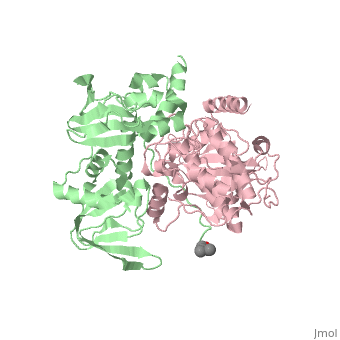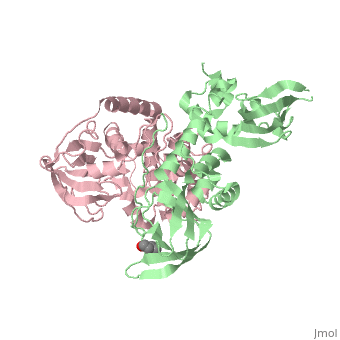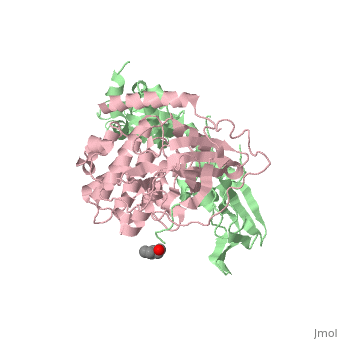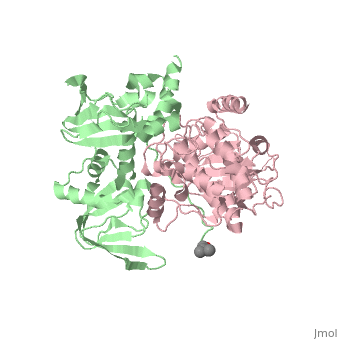We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3tnp
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | [[ | + | ==Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme== |
| + | <StructureSection load='3tnp' size='340' side='right' caption='[[3tnp]], [[Resolution|resolution]] 2.30Å' scene=''> | ||
| + | == Structural highlights == | ||
| + | <table><tr><td colspan='2'>[[3tnp]] is a 4 chain structure with sequence from [http://en.wikipedia.org/wiki/Mus_musculus Mus musculus] and [http://en.wikipedia.org/wiki/Rattus_norvegicus Rattus norvegicus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3TNP OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3TNP FirstGlance]. <br> | ||
| + | </td></tr><tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=SEP:PHOSPHOSERINE'>SEP</scene>, <scene name='pdbligand=TPO:PHOSPHOTHREONINE'>TPO</scene></td></tr> | ||
| + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3tnq|3tnq]], [[3tnr|3tnr]]</td></tr> | ||
| + | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">Pkaca, Prkaca, rCG_51506 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=10116 Rattus norvegicus]), Prkar2b ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=10090 Mus musculus])</td></tr> | ||
| + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/cAMP-dependent_protein_kinase cAMP-dependent protein kinase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.11.11 2.7.11.11] </span></td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3tnp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3tnp OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3tnp RCSB], [http://www.ebi.ac.uk/pdbsum/3tnp PDBsum]</span></td></tr> | ||
| + | </table> | ||
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/KAPCA_MOUSE KAPCA_MOUSE]] Phosphorylates a large number of substrates in the cytoplasm and the nucleus. Regulates the abundance of compartmentalized pools of its regulatory subunits through phosphorylation of PJA2 which binds and ubiquitinates these subunits, leading to their subsequent proteolysis. Phosphorylates CDC25B, ABL1, NFKB1, CLDN3, PSMC5/RPT6, PJA2, RYR2, RORA, TRPC1 and VASP. RORA is activated by phosphorylation. Required for glucose-mediated adipogenic differentiation increase and osteogenic differentiation inhibition from osteoblasts. Involved in the regulation of platelets in response to thrombin and collagen; maintains circulating platelets in a resting state by phosphorylating proteins in numerous platelet inhibitory pathways when in complex with NF-kappa-B (NFKB1 and NFKB2) and I-kappa-B-alpha (NFKBIA), but thrombin and collagen disrupt these complexes and free active PRKACA stimulates platelets and leads to platelet aggregation by phosphorylating VASP. Prevents the antiproliferative and anti-invasive effects of alpha-difluoromethylornithine in breast cancer cells when activated. RYR2 channel activity is potentiated by phosphorylation in presence of luminal Ca(2+), leading to reduced amplitude and increased frequency of store overload-induced Ca(2+) release (SOICR) characterized by an increased rate of Ca(2+) release and propagation velocity of spontaneous Ca(2+) waves, despite reduced wave amplitude and resting cytosolic Ca(2+). TRPC1 activation by phosphorylation promotes Ca(2+) influx, essential for the increase in permeability induced by thrombin in confluent endothelial monolayers. PSMC5/RPT6 activation by phosphorylation stimulates proteasome. Regulates negatively tight junction (TJs) in ovarian cancer cells via CLDN3 phosphorylation. NFKB1 phosphorylation promotes NF-kappa-B p50-p50 DNA binding. Involved in embryonic development by down-regulating the Hedgehog (Hh) signaling pathway that determines embryo pattern formation and morphogenesis. Isoform 2 phosphorylates and activates ABL1 in sperm flagellum to promote spermatozoa capacitation. Prevents meiosis resumption in prophase-arrested oocytes via CDC25B inactivation by phosphorylation. May also regulate rapid eye movement (REM) sleep in the pedunculopontine tegmental (PPT).<ref>PMID:15340140</ref> <ref>PMID:19223768</ref> <ref>PMID:19560455</ref> [[http://www.uniprot.org/uniprot/KAP3_MOUSE KAP3_MOUSE]] Regulatory subunit of the cAMP-dependent protein kinases involved in cAMP signaling in cells. Type II regulatory chains mediate membrane association by binding to anchoring proteins, including the MAP2 kinase. | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | In its physiological state, cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) is a tetramer that contains a regulatory (R) subunit dimer and two catalytic (C) subunits. We describe here the 2.3 angstrom structure of full-length tetrameric RIIbeta(2):C(2) holoenzyme. This structure showing a dimer of dimers provides a mechanistic understanding of allosteric activation by cAMP. The heterodimers are anchored together by an interface created by the beta4-beta5 loop in the RIIbeta subunit, which docks onto the carboxyl-terminal tail of the adjacent C subunit, thereby forcing the C subunit into a fully closed conformation in the absence of nucleotide. Diffusion of magnesium adenosine triphosphate (ATP) into these crystals trapped not ATP, but the reaction products, adenosine diphosphate and the phosphorylated RIIbeta subunit. This complex has implications for the dissociation-reassociation cycling of PKA. The quaternary structure of the RIIbeta tetramer differs appreciably from our model of the RIalpha tetramer, confirming the small-angle x-ray scattering prediction that the structures of each PKA tetramer are different. | ||
| - | + | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme.,Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS Science. 2012 Feb 10;335(6069):712-6. PMID:22323819<ref>PMID:22323819</ref> | |
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| + | </div> | ||
| - | + | ==See Also== | |
| - | + | *[[CAMP-dependent protein kinase|CAMP-dependent protein kinase]] | |
| - | == | + | == References == |
| - | [[ | + | <references/> |
| + | __TOC__ | ||
| + | </StructureSection> | ||
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Rattus norvegicus]] | [[Category: Rattus norvegicus]] | ||
[[Category: CAMP-dependent protein kinase]] | [[Category: CAMP-dependent protein kinase]] | ||
| - | [[Category: Deal, M S | + | [[Category: Deal, M S]] |
| - | [[Category: Keshwani, M M | + | [[Category: Keshwani, M M]] |
| - | [[Category: Kornev, A P | + | [[Category: Kornev, A P]] |
| - | [[Category: Smith-Nguyen, E V | + | [[Category: Smith-Nguyen, E V]] |
| - | [[Category: Taylor, S S | + | [[Category: Taylor, S S]] |
| - | [[Category: Zhang, P | + | [[Category: Zhang, P]] |
[[Category: Pka riib tetrameric holoenzyme]] | [[Category: Pka riib tetrameric holoenzyme]] | ||
[[Category: Transferase]] | [[Category: Transferase]] | ||
Revision as of 04:16, 25 December 2014
Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme
| |||||||||||