2axi
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:2axi.gif|left|200px]] | + | [[Image:2axi.gif|left|200px]] |
| - | + | ||
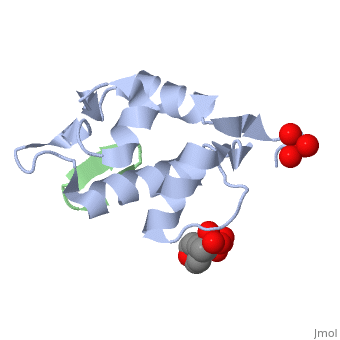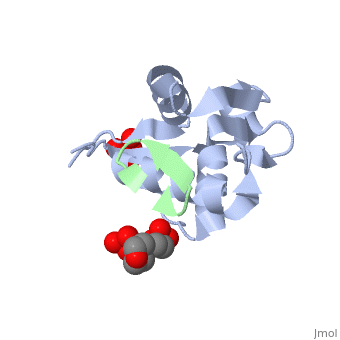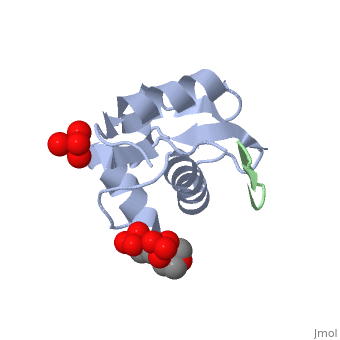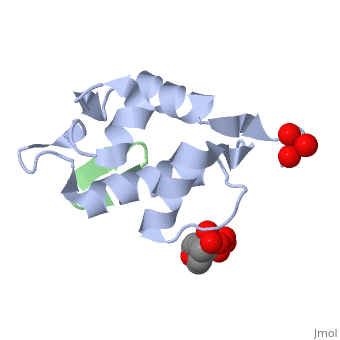
| - | '''HDM2 in complex with a beta-hairpin''' | + | {{Structure |
| + | |PDB= 2axi |SIZE=350|CAPTION= <scene name='initialview01'>2axi</scene>, resolution 1.40Å | ||
| + | |SITE= | ||
| + | |LIGAND= <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene> and <scene name='pdbligand=MPO:3[N-MORPHOLINO]PROPANE SULFONIC ACID'>MPO</scene> | ||
| + | |ACTIVITY= | ||
| + | |GENE= MDM2 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]) | ||
| + | }} | ||
| + | |||
| + | '''HDM2 in complex with a beta-hairpin''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
| - | 2AXI is a [ | + | 2AXI is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2AXI OCA]. |
==Reference== | ==Reference== | ||
| - | Structure-activity studies in a family of beta-hairpin protein epitope mimetic inhibitors of the p53-HDM2 protein-protein interaction., Fasan R, Dias RL, Moehle K, Zerbe O, Obrecht D, Mittl PR, Grutter MG, Robinson JA, Chembiochem. 2006 Mar;7(3):515-26. PMID:[http:// | + | Structure-activity studies in a family of beta-hairpin protein epitope mimetic inhibitors of the p53-HDM2 protein-protein interaction., Fasan R, Dias RL, Moehle K, Zerbe O, Obrecht D, Mittl PR, Grutter MG, Robinson JA, Chembiochem. 2006 Mar;7(3):515-26. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/16511824 16511824] |
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
| Line 21: | Line 30: | ||
[[Category: drug design]] | [[Category: drug design]] | ||
[[Category: p53]] | [[Category: p53]] | ||
| - | [[Category: protein-protein | + | [[Category: protein-protein interaction]] |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 15:55:01 2008'' |
Revision as of 13:55, 20 March 2008
| |||||||
| , resolution 1.40Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | and | ||||||
| Gene: | MDM2 (Homo sapiens) | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
HDM2 in complex with a beta-hairpin
Overview
Inhibitors of the interaction between the p53 tumor-suppressor protein and its natural human inhibitor HDM2 are attractive as potential anticancer agents. In earlier work we explored designing beta-hairpin peptidomimetics of the alpha-helical epitope on p53 that would bind tightly to the p53-binding site on HDM2. The beta-hairpin is used as a scaffold to display energetically hot residues in an optimal array for interaction with HDM2. The initial lead beta-hairpin mimetic, with a weak inhibitory activity (IC(50)=125 microM), was optimized to afford cyclo-(L-Pro-Phe-Glu-6ClTrp-Leu-Asp-Trp-Glu-Phe-D-Pro) (where 6ClTrp=L-6-chlorotryptophan), which has an affinity almost 1,000 times higher (IC(50)=140 nM). In this work, insights into the origins of this affinity maturation based on structure-activity studies and an X-ray crystal structure of the inhibitor/HDM2(residues 17-125) complex at 1.4 A resolution are described. The crystal structure confirms the beta-hairpin conformation of the bound ligand, and also reveals that a significant component of the affinity increase arises through new aromatic/aromatic stacking interactions between side chains around the hairpin and groups on the surface of HDM2.
About this Structure
2AXI is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Structure-activity studies in a family of beta-hairpin protein epitope mimetic inhibitors of the p53-HDM2 protein-protein interaction., Fasan R, Dias RL, Moehle K, Zerbe O, Obrecht D, Mittl PR, Grutter MG, Robinson JA, Chembiochem. 2006 Mar;7(3):515-26. PMID:16511824
Page seeded by OCA on Thu Mar 20 15:55:01 2008