This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Cyclooxygenase
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='5cox' size='450' side='right' caption='' > | |
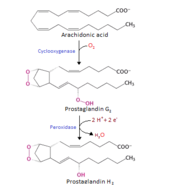
'''Cyclooxygenase COX-1''' and '''COX-2''', also called '''Prostaglandin H2 synthase PGHS-1''' and '''PGHS-2''', regulate a key step in prostaglandin and thromboxane synthesis and are the targets of nonsteroidal antiinflammatory drugs (NSAIDs) <ref name="Smith&Langenbach2001">PMID: 11413152</ref> <ref name="Chandrasekharan2002">PMID: 12242329</ref> <ref name="Ghosh2010">PMID: 20508278</ref>. Prostaglandins are implicated in various pathophysiological processes such as inflammatory reactions, gastrointestinal cytoprotection, hemostasis and thrombosis, as well as renal hemodynamics <ref name="Smith&Langenbach2001" /> <ref name="Ghosh2010"/> <ref name="Smith2000">PMID: 10966456</ref>. Whereas COX-1 presents a widespread constitutive expression, COX-2 is undetectable in most normal tissues (except for the central nervous system, kidneys, and seminal vesicles), but is induced by various inflammatory and mitogenic stimuli <ref name="Smith2000" /> <ref name="Ghosh2010"/> <ref name="Rang&Dale2008">Rang HP, Dale MM, Ritter JM, Flower RJ. 2008. Pharmacology. Elsevier. 6th edition. UK. 844 p.</ref>. More recently, a third isoform named COX-3 was identified as a COX-1 splicing variant. This new isoform may play a role in processes such as fever and pain <ref name="Ghosh2010"/> <ref name="Chandrasekharan2002"/>. | '''Cyclooxygenase COX-1''' and '''COX-2''', also called '''Prostaglandin H2 synthase PGHS-1''' and '''PGHS-2''', regulate a key step in prostaglandin and thromboxane synthesis and are the targets of nonsteroidal antiinflammatory drugs (NSAIDs) <ref name="Smith&Langenbach2001">PMID: 11413152</ref> <ref name="Chandrasekharan2002">PMID: 12242329</ref> <ref name="Ghosh2010">PMID: 20508278</ref>. Prostaglandins are implicated in various pathophysiological processes such as inflammatory reactions, gastrointestinal cytoprotection, hemostasis and thrombosis, as well as renal hemodynamics <ref name="Smith&Langenbach2001" /> <ref name="Ghosh2010"/> <ref name="Smith2000">PMID: 10966456</ref>. Whereas COX-1 presents a widespread constitutive expression, COX-2 is undetectable in most normal tissues (except for the central nervous system, kidneys, and seminal vesicles), but is induced by various inflammatory and mitogenic stimuli <ref name="Smith2000" /> <ref name="Ghosh2010"/> <ref name="Rang&Dale2008">Rang HP, Dale MM, Ritter JM, Flower RJ. 2008. Pharmacology. Elsevier. 6th edition. UK. 844 p.</ref>. More recently, a third isoform named COX-3 was identified as a COX-1 splicing variant. This new isoform may play a role in processes such as fever and pain <ref name="Ghosh2010"/> <ref name="Chandrasekharan2002"/>. | ||
| Line 19: | Line 19: | ||
==Structure <ref name="Garavito2003>PMID: 17851687</ref> <ref name="Smith2000"/>== | ==Structure <ref name="Garavito2003>PMID: 17851687</ref> <ref name="Smith2000"/>== | ||
| - | + | ||
In 1994, Picot ''et al'' published the first three-dimensional (3D) structure of a COX enzyme, the ovine COX-1 complexed with the NSAID flurbiprofen. Soon afterward, the crystal structures of human and murine COX-2 followed. First, the three-dimensional structure of human COX-2 was assessed by means of sequence homology modeling, but in 1996, Luong, C. ''et al'' <ref name="Luong1996">PMID: 8901870</ref> and Kurumbail, R.G ''et al'' <ref name="Kurumbail1996">PMID: 8967954</ref> published two crystal structures of the recombinant human and mouse COX-2 isozymes, respectively, complexed with different selective inhibitors. Given its pharmacological importance as a therapeutic target, drug interactions with COX were one of the first issues to be addressed, and complexes containing a number of different NSAIDs have been studied crystallographically. The structural analysis of COX complexed with substrates or products was more difficult to pursue for a number of technical reasons. However, within the past years, crystal structures of murine COX-2 complexed with AA and EPA have also been determined. | In 1994, Picot ''et al'' published the first three-dimensional (3D) structure of a COX enzyme, the ovine COX-1 complexed with the NSAID flurbiprofen. Soon afterward, the crystal structures of human and murine COX-2 followed. First, the three-dimensional structure of human COX-2 was assessed by means of sequence homology modeling, but in 1996, Luong, C. ''et al'' <ref name="Luong1996">PMID: 8901870</ref> and Kurumbail, R.G ''et al'' <ref name="Kurumbail1996">PMID: 8967954</ref> published two crystal structures of the recombinant human and mouse COX-2 isozymes, respectively, complexed with different selective inhibitors. Given its pharmacological importance as a therapeutic target, drug interactions with COX were one of the first issues to be addressed, and complexes containing a number of different NSAIDs have been studied crystallographically. The structural analysis of COX complexed with substrates or products was more difficult to pursue for a number of technical reasons. However, within the past years, crystal structures of murine COX-2 complexed with AA and EPA have also been determined. | ||
| Line 48: | Line 48: | ||
=====Cyclooxygenase Active Site Structure===== | =====Cyclooxygenase Active Site Structure===== | ||
PGHS-1 and 2 monomers each contain a 25-°A hydrophobic channel that originates at the membrane binding domain and extends into the core of the globular domain. The MBD forms the entrance and the first half of the channel and allows arachidonate and O2 to enter directly from the apolar compartment of the lipid bilayer. Several amino acids composing the upper half of the channel are uniquely important to cyclooxygenase catalysis. Twenty-four residues line the hydrophobic <scene name='SandboxUAM/Mynewscene/25'>cyclooxygenase active site</scene> with only one difference between the isozymes—Ile at position 523 in PGHS-1 and Val at position 523 in PGHS-2. Amino acids lining the hydrophobic cyclooxygenase active site channel include Leu117, Arg120, Phe205, Phe209, Val344, Ile345, Tyr348, Val349, Leu352, Ser353, Tyr355, Leu359, Phe381, Leu384, Tyr385, Trp387, Phe518, Ile/Val523, Gly526, Ala527, Ser530, Leu531, Gly533, Leu534. <scene name='SandboxUAM/Mynewscene/18'>Only three of the channel residues</scene> are polar (Arg120, Ser353, and Ser530).<scene name='SandboxUAM/Mynewscene/26'>Tyr 385</scene> in its radical form is the responsible for abstracting a proton from arachidonic acid during its conversion to PGG2.<scene name='SandboxUAM/Mynewscene/27'>Ser530</scene> is the site of acetylation by [[Aspirin effects on COX aka PGHS|aspirin]] and <scene name='SandboxUAM/Mynewscene/30'>Arg120</scene>, which is positioned about midway between the entrance and the apex of the active site <ref name="Garavito&DeWitt1999">PMID: 10570255</ref>, binds to the carboxylate groups of fatty acids and many NSAIDs. <br/> | PGHS-1 and 2 monomers each contain a 25-°A hydrophobic channel that originates at the membrane binding domain and extends into the core of the globular domain. The MBD forms the entrance and the first half of the channel and allows arachidonate and O2 to enter directly from the apolar compartment of the lipid bilayer. Several amino acids composing the upper half of the channel are uniquely important to cyclooxygenase catalysis. Twenty-four residues line the hydrophobic <scene name='SandboxUAM/Mynewscene/25'>cyclooxygenase active site</scene> with only one difference between the isozymes—Ile at position 523 in PGHS-1 and Val at position 523 in PGHS-2. Amino acids lining the hydrophobic cyclooxygenase active site channel include Leu117, Arg120, Phe205, Phe209, Val344, Ile345, Tyr348, Val349, Leu352, Ser353, Tyr355, Leu359, Phe381, Leu384, Tyr385, Trp387, Phe518, Ile/Val523, Gly526, Ala527, Ser530, Leu531, Gly533, Leu534. <scene name='SandboxUAM/Mynewscene/18'>Only three of the channel residues</scene> are polar (Arg120, Ser353, and Ser530).<scene name='SandboxUAM/Mynewscene/26'>Tyr 385</scene> in its radical form is the responsible for abstracting a proton from arachidonic acid during its conversion to PGG2.<scene name='SandboxUAM/Mynewscene/27'>Ser530</scene> is the site of acetylation by [[Aspirin effects on COX aka PGHS|aspirin]] and <scene name='SandboxUAM/Mynewscene/30'>Arg120</scene>, which is positioned about midway between the entrance and the apex of the active site <ref name="Garavito&DeWitt1999">PMID: 10570255</ref>, binds to the carboxylate groups of fatty acids and many NSAIDs. <br/> | ||
| - | </StructureSection> | ||
| - | |||
==NSAIDs== | ==NSAIDs== | ||
| - | <Structure load='3ln1' size='300' frame='true' align='right' caption='COX-2 bound to celecoxib [[3ln1]]' scene='Insert optional scene name here' /> | ||
| - | |||
Non-steroid anti-inflammatory [[Pharma drugs|drugs]] are a chemically heterogeneous group of compounds whose major function is the inhibition of cyclooxygenases (Table 1). Apart from their anti-inflammatory effect, they also present analgesic and antipyretic properties <ref name="Rang&Dale2008"/>. | Non-steroid anti-inflammatory [[Pharma drugs|drugs]] are a chemically heterogeneous group of compounds whose major function is the inhibition of cyclooxygenases (Table 1). Apart from their anti-inflammatory effect, they also present analgesic and antipyretic properties <ref name="Rang&Dale2008"/>. | ||
| Line 129: | Line 125: | ||
Finally, paracetamol is considered an atypical NSAIDs, not only because of its lack of anti-inflammatory properties but also because it does not interact neither with COX-1 nor with COX-2 <ref name="Rang&Dale2008"/>. It has been proposed that paracetamol may act as an analgesic and antipyretic drug by inhibition of COX-3 <ref name="Rang&Dale2008"/>. | Finally, paracetamol is considered an atypical NSAIDs, not only because of its lack of anti-inflammatory properties but also because it does not interact neither with COX-1 nor with COX-2 <ref name="Rang&Dale2008"/>. It has been proposed that paracetamol may act as an analgesic and antipyretic drug by inhibition of COX-3 <ref name="Rang&Dale2008"/>. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Regulation== | ==Regulation== | ||
| Line 179: | Line 161: | ||
* For more information about inflammation, see: [[Inflammation]] | * For more information about inflammation, see: [[Inflammation]] | ||
* For information about other Pharmaceutical Therapeutics, see: [[Pharmaceutical Therapeutics]] | * For information about other Pharmaceutical Therapeutics, see: [[Pharmaceutical Therapeutics]] | ||
| - | + | </StructureSection> | |
| + | __NOTOC__ | ||
== 3D Structures of cyclooxygenase == | == 3D Structures of cyclooxygenase == | ||
Revision as of 09:18, 10 July 2013
| |||||||||||
3D Structures of cyclooxygenase
Updated on 10-July-2013
COX I
3n8v - sCOX I – sheep
1pth – sCOX I + aspirin
1pge, 1pgf, 1pgg - sCOX I + iodinated anti-inflammatory drug
1cqe, 3kk6 - sCOX I + anti-inflammatory inhibitor
1igx, 1igz - sCOX I + linoleic acid derivative
1u67 - sCOX I (mutant) + fatty acid substrate
2ayl - sCOX I Mn-substituted + anti-inflammatory drug
3n8w - sCOX I (mutant) + anti-inflammatory drug
3n8x, 3n8z - sCOX I + anti-inflammatory drug
3n8y - sCOX I + aspirin + anti-inflammatory drug
2oye, 2oyu - sCOX I + inhibitor
COX II
5cox - mCOX II]] - mouse
1cx2, 3pgh, 4cox, 6cox - mCOX II + anti-inflammatory inhibitor
1pxx, 3ln1, 3q7d, 3rr3 - mCOX II + anti-inflammatory drug
1q4g - mCOX II + inhibitor
3qmo - mCOX II (mutant) + inhibitor
3hs5, 3hs6, 3hs7, 3krk, 3mdl, 3olt, 3olu, 3qh0 - mCOX II (mutant) + fatty acid substrate
3ln0, 3mqe, 3ntg - mCOX II + benzopyran inhibitor
Authorship
This page has been constructed and edited by Maria Saiz, Rafael Gonzalez & Eva Garcia, students of the Biomedicine Master of the Universidad Autonoma de Madrid (Spain) under the supervision of Dr. Cristina Murga, as a contribution to the Molecular Pharmacology Course of the doctoral programme in Biosciences.
References
- ↑ 1.0 1.1 Smith WL, Langenbach R. Why there are two cyclooxygenase isozymes. J Clin Invest. 2001 Jun;107(12):1491-5. PMID:11413152 doi:10.1172/JCI13271
- ↑ 2.0 2.1 Chandrasekharan NV, Dai H, Roos KL, Evanson NK, Tomsik J, Elton TS, Simmons DL. COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: cloning, structure, and expression. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13926-31. Epub 2002 Sep 19. PMID:12242329 doi:10.1073/pnas.162468699
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 Ghosh N, Chaki R, Mandal V, Mandal SC. COX-2 as a target for cancer chemotherapy. Pharmacol Rep. 2010 Mar-Apr;62(2):233-44. PMID:20508278
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145-82. PMID:10966456 doi:10.1146/annurev.biochem.69.1.145
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 Rang HP, Dale MM, Ritter JM, Flower RJ. 2008. Pharmacology. Elsevier. 6th edition. UK. 844 p.
- ↑ Barnes NL, Warnberg F, Farnie G, White D, Jiang W, Anderson E, Bundred NJ. Cyclooxygenase-2 inhibition: effects on tumour growth, cell cycling and lymphangiogenesis in a xenograft model of breast cancer. Br J Cancer. 2007 Feb 26;96(4):575-82. Epub 2007 Feb 6. PMID:17285134 doi:10.1038/sj.bjc.6603593
- ↑ Boland GP, Butt IS, Prasad R, Knox WF, Bundred NJ. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer. 2004 Jan 26;90(2):423-9. PMID:14735188 doi:10.1038/sj.bjc.6601534
- ↑ Sales KJ, Grant V, Jabbour HN. Prostaglandin E2 and F2alpha activate the FP receptor and up-regulate cyclooxygenase-2 expression via the cyclic AMP response element. Mol Cell Endocrinol. 2008 Mar 26;285(1-2):51-61. Epub 2008 Feb 3. PMID:18316157 doi:10.1016/j.mce.2008.01.016
- ↑ Perrone G, Zagami M, Altomare V, Battista C, Morini S, Rabitti C. COX-2 localization within plasma membrane caveolae-like structures in human lobular intraepithelial neoplasia of the breast. Virchows Arch. 2007 Dec;451(6):1039-45. Epub 2007 Sep 13. PMID:17851687 doi:10.1007/s00428-007-0506-4
- ↑ Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner MF. Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat Struct Biol. 1996 Nov;3(11):927-33. PMID:8901870
- ↑ Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996 Dec 19-26;384(6610):644-8. PMID:8967954 doi:http://dx.doi.org/10.1038/384644a0
- ↑ Spencer AG, Thuresson E, Otto JC, Song I, Smith T, DeWitt DL, Garavito RM, Smith WL. The membrane binding domains of prostaglandin endoperoxide H synthases 1 and 2. Peptide mapping and mutational analysis. J Biol Chem. 1999 Nov 12;274(46):32936-42. PMID:10551860
- ↑ Vecchio AJ, Simmons DM, Malkowski MG. Structural basis of fatty acid substrate binding to cyclooxygenase-2. J Biol Chem. 2010 Jul 16;285(29):22152-63. Epub 2010 May 12. PMID:20463020 doi:10.1074/jbc.M110.119867
- ↑ Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner MF. Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat Struct Biol. 1996 Nov;3(11):927-33. PMID:8901870
- ↑ 15.0 15.1 15.2 Garavito RM, DeWitt DL. The cyclooxygenase isoforms: structural insights into the conversion of arachidonic acid to prostaglandins. Biochim Biophys Acta. 1999 Nov 23;1441(2-3):278-87. PMID:10570255
- ↑ FitzGerald GA, Loll P. COX in a crystal ball: current status and future promise of prostaglandin research. J Clin Invest. 2001 Jun;107(11):1335-7. PMID:11390412 doi:10.1172/JCI13037
- ↑ Harper KA, Tyson-Capper AJ. Complexity of COX-2 gene regulation. Biochem Soc Trans. 2008 Jun;36(Pt 3):543-5. PMID:18482003 doi:10.1042/BST0360543
- ↑ Deng WG, Zhu Y, Wu KK. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J Biol Chem. 2003 Feb 14;278(7):4770-7. Epub 2002 Dec 5. PMID:12471036 doi:10.1074/jbc.M209286200
- ↑ Song SH, Jong HS, Choi HH, Inoue H, Tanabe T, Kim NK, Bang YJ. Transcriptional silencing of Cyclooxygenase-2 by hyper-methylation of the 5' CpG island in human gastric carcinoma cells. Cancer Res. 2001 Jun 1;61(11):4628-35. PMID:11389100
- ↑ Tyson-Capper AJ, Cork DM, Wesley E, Shiells EA, Loughney AD. Characterization of cellular retinoid-binding proteins in human myometrium during pregnancy. Mol Hum Reprod. 2006 Nov;12(11):695-701. Epub 2006 Sep 7. PMID:16959971 doi:10.1093/molehr/gal070
- ↑ 21.0 21.1 Yamaguchi K, Lantowski A, Dannenberg AJ, Subbaramaiah K. Histone deacetylase inhibitors suppress the induction of c-Jun and its target genes including COX-2. J Biol Chem. 2005 Sep 23;280(38):32569-77. Epub 2005 Jul 1. PMID:15994313 doi:10.1074/jbc.M503201200
- ↑ Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3'-untranslated region. J Biol Chem. 2000 Apr 21;275(16):11750-7. PMID:10766797
- ↑ Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003 Jan;11(1):113-26. PMID:12535526
- ↑ Subbaramaiah K, Marmo TP, Dixon DA, Dannenberg AJ. Regulation of cyclooxgenase-2 mRNA stability by taxanes: evidence for involvement of p38, MAPKAPK-2, and HuR. J Biol Chem. 2003 Sep 26;278(39):37637-47. Epub 2003 Jun 25. PMID:12826679 doi:10.1074/jbc.M301481200
- ↑ Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005 May 4;33(8):2565-79. Print 2005. PMID:15872218 doi:10.1093/nar/gki544
- ↑ Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR. RNA. 2007 Jul;13(7):1103-15. Epub 2007 May 16. PMID:17507659 doi:10.1261/rna.577707
Proteopedia Page Contributors and Editors (what is this?)
Cristina Murga, Michal Harel, David Canner, María Laura Saiz Álvarez, Alexander Berchansky