This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2pol
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ==THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP== | |
| - | + | <StructureSection load='2pol' size='340' side='right' caption='[[2pol]], [[Resolution|resolution]] 2.50Å' scene=''> | |
| - | + | == Structural highlights == | |
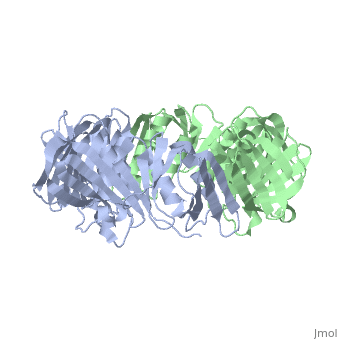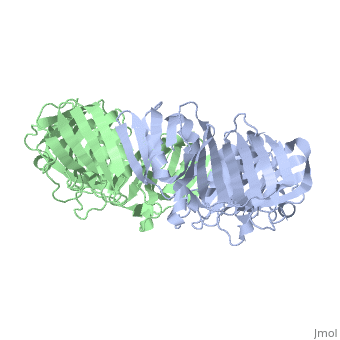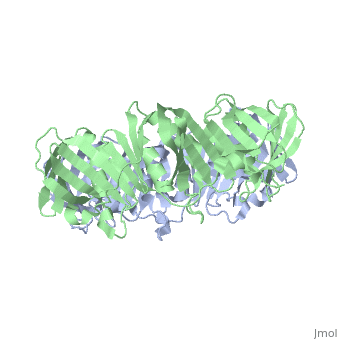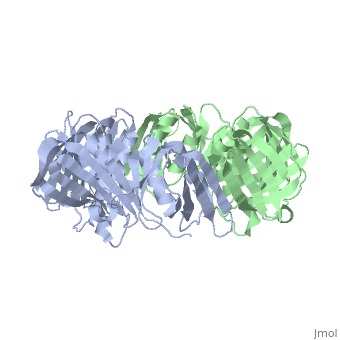
| + | <table><tr><td colspan='2'>[[2pol]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. The June 2012 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Sliding Clamps'' by David Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2012_6 10.2210/rcsb_pdb/mom_2012_6]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2POL OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2POL FirstGlance]. <br> | ||
| + | </td></tr><tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">POL ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli])</td></tr> | ||
| + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/DNA-directed_DNA_polymerase DNA-directed DNA polymerase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.7.7 2.7.7.7] </span></td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2pol FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pol OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2pol RCSB], [http://www.ebi.ac.uk/pdbsum/2pol PDBsum]</span></td></tr> | ||
| + | </table> | ||
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/DPO3B_ECOLI DPO3B_ECOLI]] DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity. The beta chain is required for initiation of replication once it is clamped onto DNA, it slides freely (bidirectional and ATP-independent) along duplex DNA. | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/po/2pol_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The crystal structure of the beta subunit (processivity factor) of DNA polymerase III holoenzyme has been determined at 2.5 A resolution. A dimer of the beta subunit (M(r) = 2 x 40.6 kd, 2 x 366 amino acid residues) forms a ring-shaped structure lined by 12 alpha helices that can encircle duplex DNA. The structure is highly symmetrical, with each monomer containing three domains of identical topology. The charge distribution and orientation of the helices indicate that the molecule functions by forming a tight clamp that can slide on DNA, as shown biochemically. A potential structural relationship is suggested between the beta subunit and proliferating cell nuclear antigen (PCNA, the eukaryotic polymerase delta [and epsilon] processivity factor), and the gene 45 protein of the bacteriophage T4 DNA polymerase. | ||
| - | + | Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp.,Kong XP, Onrust R, O'Donnell M, Kuriyan J Cell. 1992 May 1;69(3):425-37. PMID:1349852<ref>PMID:1349852</ref> | |
| - | + | ||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
==See Also== | ==See Also== | ||
*[[DNA polymerase|DNA polymerase]] | *[[DNA polymerase|DNA polymerase]] | ||
*[[Polymerase III homoenzyme beta subunit|Polymerase III homoenzyme beta subunit]] | *[[Polymerase III homoenzyme beta subunit|Polymerase III homoenzyme beta subunit]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: DNA-directed DNA polymerase]] | [[Category: DNA-directed DNA polymerase]] | ||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Sliding Clamps]] | [[Category: Sliding Clamps]] | ||
| - | [[Category: Kong, X P | + | [[Category: Kong, X P]] |
| - | [[Category: Kuriyan, J | + | [[Category: Kuriyan, J]] |
[[Category: Nucleotidyltransferase]] | [[Category: Nucleotidyltransferase]] | ||
Revision as of 04:49, 25 December 2014
THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP
| |||||||||||