We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1jch
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
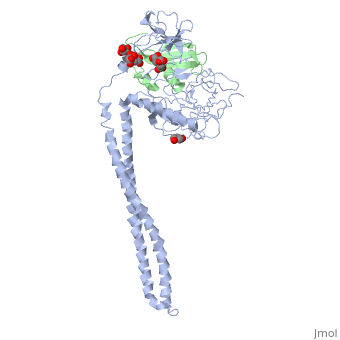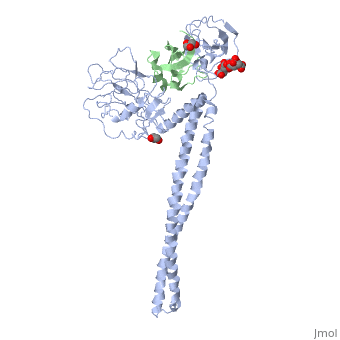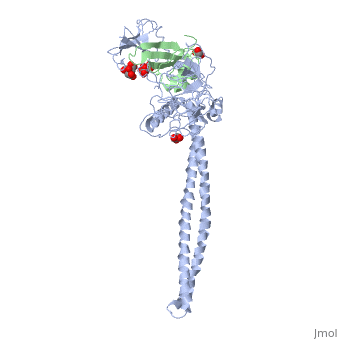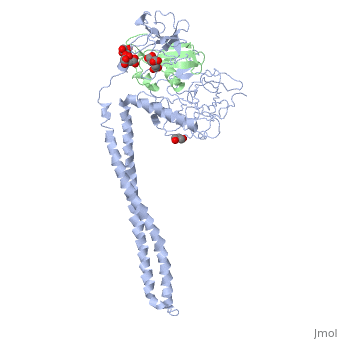
| - | + | ==Crystal Structure of Colicin E3 in Complex with its Immunity Protein== | |
| - | + | <StructureSection load='1jch' size='340' side='right' caption='[[1jch]], [[Resolution|resolution]] 3.02Å' scene=''> | |
| - | + | == Structural highlights == | |
| + | <table><tr><td colspan='2'>[[1jch]] is a 4 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli_str._k-12_substr._w3110 Escherichia coli str. k-12 substr. w3110]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JCH OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1JCH FirstGlance]. <br> | ||
| + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CIT:CITRIC+ACID'>CIT</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene></td></tr> | ||
| + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3eip|3eip]], [[1e44|1e44]]</td></tr> | ||
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1jch FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1jch OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1jch RCSB], [http://www.ebi.ac.uk/pdbsum/1jch PDBsum]</span></td></tr> | ||
| + | </table> | ||
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/CEA3_ECOLX CEA3_ECOLX]] Inactivates ribosomes by hydrolyzing 16S RNA in 30S ribosomes at a specific site. Colicins are polypeptide toxins produced by and active against E.coli and closely related bacteria. [[http://www.uniprot.org/uniprot/IMM3_ECOLX IMM3_ECOLX]] This protein inhibits the 16S RNA hydrolyzing activity of colicin E3 by binding with high affinity to the C-terminal catalytic domain of E3. This protein is able to protect a cell, which harbors the plasmid ColE3 against colicin E3. | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/jc/1jch_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Colicins kill E. coli by a process that involves binding to a surface receptor, entering the cell, and, finally, intoxicating it. The lethal action of colicin E3 is a specific cleavage in the ribosomal decoding A site. The crystal structure of colicin E3, reported here in a binary complex with its immunity protein (IP), reveals a Y-shaped molecule with the receptor binding domain forming a 100 A long stalk and the two globular heads of the translocation domain (T) and the catalytic domain (C) comprising the two arms. Active site residues are D510, H513, E517, and R545. IP is buried between T and C. Rather than blocking the active site, IP prevents access of the active site to the ribosome. | ||
| - | + | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.,Soelaiman S, Jakes K, Wu N, Li C, Shoham M Mol Cell. 2001 Nov;8(5):1053-62. PMID:11741540<ref>PMID:11741540</ref> | |
| - | + | ||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
==See Also== | ==See Also== | ||
| Line 10: | Line 32: | ||
*[[Colicin Immunity Protein|Colicin Immunity Protein]] | *[[Colicin Immunity Protein|Colicin Immunity Protein]] | ||
*[[Im3|Im3]] | *[[Im3|Im3]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Escherichia coli str. k-12 substr. w3110]] | [[Category: Escherichia coli str. k-12 substr. w3110]] | ||
| - | [[Category: Jakes, K | + | [[Category: Jakes, K]] |
| - | [[Category: Li, C | + | [[Category: Li, C]] |
| - | [[Category: Shoham, M | + | [[Category: Shoham, M]] |
| - | [[Category: Soelaiman, S | + | [[Category: Soelaiman, S]] |
| - | [[Category: Wu, N | + | [[Category: Wu, N]] |
[[Category: Hydrolase]] | [[Category: Hydrolase]] | ||
[[Category: Ribosome inhibitor]] | [[Category: Ribosome inhibitor]] | ||
Revision as of 21:34, 25 December 2014
Crystal Structure of Colicin E3 in Complex with its Immunity Protein
| |||||||||||
Categories: Escherichia coli str. k-12 substr. w3110 | Jakes, K | Li, C | Shoham, M | Soelaiman, S | Wu, N | Hydrolase | Ribosome inhibitor | The receptor-binding domain is a coiled coil | The rnase domain is a six-stranded antiparallel beta-sheet. the immunity protein is a four-stranded antiparallel beta sheet flanked by 3 helices on one side of the sheet | Translocation domain is a beta-jellyroll