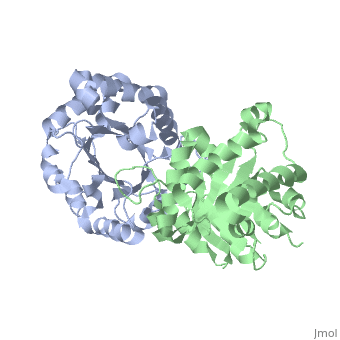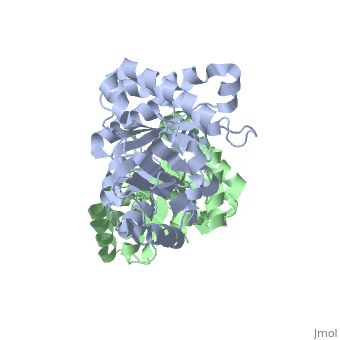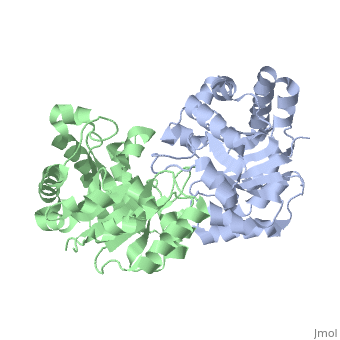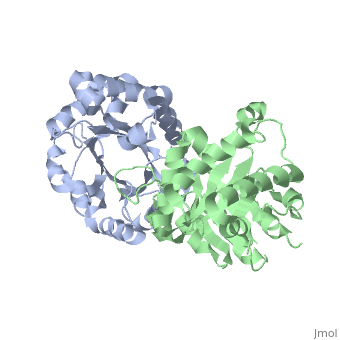
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1ypi
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ==STRUCTURE OF YEAST TRIOSEPHOSPHATE ISOMERASE AT 1.9-ANGSTROMS RESOLUTION== | |
| - | + | <StructureSection load='1ypi' size='340' side='right' caption='[[1ypi]], [[Resolution|resolution]] 1.90Å' scene=''> | |
| - | + | == Structural highlights == | |
| + | <table><tr><td colspan='2'>[[1ypi]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1YPI OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1YPI FirstGlance]. <br> | ||
| + | </td></tr><tr><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Triose-phosphate_isomerase Triose-phosphate isomerase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=5.3.1.1 5.3.1.1] </span></td></tr> | ||
| + | <tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ypi FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ypi OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1ypi RCSB], [http://www.ebi.ac.uk/pdbsum/1ypi PDBsum]</span></td></tr> | ||
| + | <table> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/yp/1ypi_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The structure of yeast triosephosphate isomerase (TIM) has been solved at 3.0-A resolution and refined at 1.9-A resolution to an R factor of 21.0%. The final model consists of all non-hydrogen atoms in the polypeptide chain and 119 water molecules, a number of which are found in the interior of the protein. The structure of the active site clearly indicates that the carboxylate of the catalytic base, Glu 165, is involved in a hydrogen-bonding interaction with the hydroxyl of Ser 96. In addition, the interactions of the other active site residues, Lys 12 and His 95, are also discussed. For the first time in any TIM structure, the "flexible loop" has well-defined density; the conformation of the loop in this structure is stabilized by a crystal contact. Analysis of the subunit interface of this dimeric enzyme hints at the source of the specificity of one subunit for another and allows us to estimate an association constant of 10(14)-10(16) M-1 for the two monomers. The analysis also suggests that the interface may be a particularly good target for drug design. The conserved positions (20%) among sequences from 13 sources ranging on the evolutionary scale from Escherichia coli to humans reveal the intense pressure to maintain the active site structure. | ||
| - | + | Structure of yeast triosephosphate isomerase at 1.9-A resolution.,Lolis E, Alber T, Davenport RC, Rose D, Hartman FC, Petsko GA Biochemistry. 1990 Jul 17;29(28):6609-18. PMID:2204417<ref>PMID:2204417</ref> | |
| - | + | ||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
==See Also== | ==See Also== | ||
*[[Triose Phosphate Isomerase|Triose Phosphate Isomerase]] | *[[Triose Phosphate Isomerase|Triose Phosphate Isomerase]] | ||
| - | + | == References == | |
| - | == | + | <references/> |
| - | < | + | __TOC__ |
| + | </StructureSection> | ||
[[Category: Saccharomyces cerevisiae]] | [[Category: Saccharomyces cerevisiae]] | ||
[[Category: Triose-phosphate isomerase]] | [[Category: Triose-phosphate isomerase]] | ||
Revision as of 21:44, 29 September 2014
STRUCTURE OF YEAST TRIOSEPHOSPHATE ISOMERASE AT 1.9-ANGSTROMS RESOLUTION
| |||||||||||