Caspase-3 Regulatory Mechanisms
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='2h5i_mm1-1.pdb' size='300' side='right' caption='Caspase-3 (PDB entry [[2h5i]])' scene='Sandox_Bay_Serrano/Monomer/1'> | ||
| + | |||
== Introduction == | == Introduction == | ||
| Line 5: | Line 7: | ||
Any apoptotic signal received by the cell causes the activation of initiator caspases (-8 and -9) by associating with other protein platforms to form a functional holoenzyme. These initiator caspases then cleave the executioner caspases -3, -6, and -7. Caspase-3 specifically functions to cleave downstream apoptotic targets as well as both caspase-6 and -7, which in turn cleave their respective targets to induce cell death. Aside from being able to activate caspase-6 and -7, caspase-3 also regulates caspase-9 activity, operating via a feedback loop. This dual action of caspase-3 confers its distinct regulatory mechanisms, resulting in a wider extent of its effects in the apoptotic cascade. | Any apoptotic signal received by the cell causes the activation of initiator caspases (-8 and -9) by associating with other protein platforms to form a functional holoenzyme. These initiator caspases then cleave the executioner caspases -3, -6, and -7. Caspase-3 specifically functions to cleave downstream apoptotic targets as well as both caspase-6 and -7, which in turn cleave their respective targets to induce cell death. Aside from being able to activate caspase-6 and -7, caspase-3 also regulates caspase-9 activity, operating via a feedback loop. This dual action of caspase-3 confers its distinct regulatory mechanisms, resulting in a wider extent of its effects in the apoptotic cascade. | ||
| - | |||
| - | ---- | ||
== Overview of Caspase-3 Structure == | == Overview of Caspase-3 Structure == | ||
| - | |||
| - | <StructureSection load='2h5i_mm1-1.pdb' size='300' side='right' caption='Caspase-3 (PDB entry [[2h5i]])' scene='Sandox_Bay_Serrano/Monomer/1'> | ||
===Dimer Formation === | ===Dimer Formation === | ||
| Line 18: | Line 16: | ||
The active pocket of caspase-3 is defined by <scene name='Sandox_Bay_Serrano/Monomer/2'>Cys-163 and His-121</scene>. Binding of a <scene name='Sandox_Bay_Serrano/Scene01_substrate/4'>substrate</scene>, such as DEVD-CHO to the active site of the enzyme induces a conformational change that allows the L2 and L2' loops to interlock and stabilize the active site <scene name='Sandox_Bay_Serrano/Scene01_substrate/3'></scene>. Like caspase-7, caspase-3 recognizes a Asp-X-X-Asp sequence as a cleavage site in its protein substrates. | The active pocket of caspase-3 is defined by <scene name='Sandox_Bay_Serrano/Monomer/2'>Cys-163 and His-121</scene>. Binding of a <scene name='Sandox_Bay_Serrano/Scene01_substrate/4'>substrate</scene>, such as DEVD-CHO to the active site of the enzyme induces a conformational change that allows the L2 and L2' loops to interlock and stabilize the active site <scene name='Sandox_Bay_Serrano/Scene01_substrate/3'></scene>. Like caspase-7, caspase-3 recognizes a Asp-X-X-Asp sequence as a cleavage site in its protein substrates. | ||
| - | ---- | ||
| - | |||
| - | </StructureSection> | ||
== Caspase-3 Loop Bundle and Active Site== | == Caspase-3 Loop Bundle and Active Site== | ||
| Line 39: | Line 34: | ||
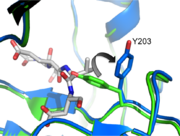
In order to be active and cleave specific apoptotic targets, Caspase-3 must be able to first bind substrate. There are several essential interactions responsible for securing the substrate before cleavage. The binding pocket at <scene name='Caspase-3_Regulatory_Mechanisms/P2/2'>P2</scene> is a hydrophobic patch made up of Y204, W206, and F250 (dark blue residues). This creates a hydrophobic pocket for the P2 residue (in this case, valine), helping it stick to the protein. At <scene name='Caspase-3_Regulatory_Mechanisms/P4/2'>P4</scene> there are contacts that contribute to the specificity of caspase-3. Asparagine 208 hydrogen bonds with an aspartate at P4 along with the backbone nitrogen of F250, creating a preference for a carboxylic acid at the P4 site. | In order to be active and cleave specific apoptotic targets, Caspase-3 must be able to first bind substrate. There are several essential interactions responsible for securing the substrate before cleavage. The binding pocket at <scene name='Caspase-3_Regulatory_Mechanisms/P2/2'>P2</scene> is a hydrophobic patch made up of Y204, W206, and F250 (dark blue residues). This creates a hydrophobic pocket for the P2 residue (in this case, valine), helping it stick to the protein. At <scene name='Caspase-3_Regulatory_Mechanisms/P4/2'>P4</scene> there are contacts that contribute to the specificity of caspase-3. Asparagine 208 hydrogen bonds with an aspartate at P4 along with the backbone nitrogen of F250, creating a preference for a carboxylic acid at the P4 site. | ||
| - | |||
| - | |||
| - | |||
| - | </StructureSection> | ||
== Caspase-3 Regulation== | == Caspase-3 Regulation== | ||
| - | |||
| - | <StructureSection load='1I3O' size='350' side='right' caption='Structure of Caspase-3 with BIR2 domain (PDB entry [[1i3o]])' scene='Caspase-3_Regulatory_Mechanisms/Bir2/1'> | ||
=== Exosite and Allosteric Site=== | === Exosite and Allosteric Site=== | ||
| Line 70: | Line 59: | ||
X-linked inhibitor of apoptosis proteins (XIAP) contains the second baculovirus IAP repeat domain (BIR2) targeting caspase-3 and caspase-7. The <scene name='Caspase-3_Regulatory_Mechanisms/Bir2/2'>BIR2</scene> domain sits directly in the active site of caspase-3 and completely inhibits the protein. The region binding the active site runs in the opposite direction of normal caspase-3 substrates, thus occupying P1 through P4 but avoiding cleavage by the protease. This unique method of inhibition is a critical regulatory mechanism used in cells to control apoptotic caspase activity. | X-linked inhibitor of apoptosis proteins (XIAP) contains the second baculovirus IAP repeat domain (BIR2) targeting caspase-3 and caspase-7. The <scene name='Caspase-3_Regulatory_Mechanisms/Bir2/2'>BIR2</scene> domain sits directly in the active site of caspase-3 and completely inhibits the protein. The region binding the active site runs in the opposite direction of normal caspase-3 substrates, thus occupying P1 through P4 but avoiding cleavage by the protease. This unique method of inhibition is a critical regulatory mechanism used in cells to control apoptotic caspase activity. | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
=References= | =References= | ||
| Line 77: | Line 68: | ||
Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466. | Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466. | ||
| - | |||
| - | </StructureSection> | ||
Revision as of 09:38, 10 July 2013
| |||||||||||
References
Bose, K., C. Pop, et al. (2003). "An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3." Biochemistry 42(42): 12298-12310.
Boucher, D., V. Blais, et al. (2012). "Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis." Proc Natl Acad Sci U S A 109(15): 5669-5674.
Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466.
Proteopedia Page Contributors and Editors (what is this?)
Scott Eron, Banyuhay P. Serrano, Alexander Berchansky, Yunlong Zhao, Jaime Prilusky, Michal Harel