HMG-CoA Reductase
From Proteopedia
| Line 3: | Line 3: | ||
|OLDID=1159684 | |OLDID=1159684 | ||
}} | }} | ||
| - | + | <StructureSection load='1dq8' size='500' side='left' scene='HMG-CoA_Reductase/1dq8_starting_scene/1' caption='Crystal Structure of HMG-CoA, [[1dq8]])'> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
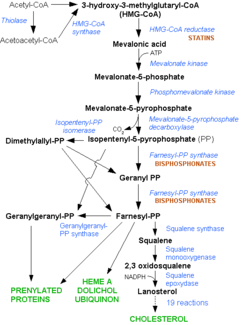
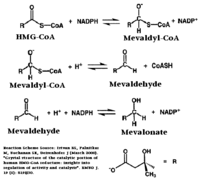
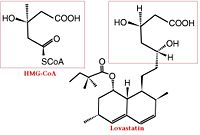
[[HMG-CoA Reductase]] (or '''3-hydroxy-3-methyl-glutaryl-CoA reductase''' or '''HMGR''') is the rate-controlling enzyme of the mevalonate pathway, responsible for cholesterol and other isoprenoid biosynthesis. HMGR is a transmembrane protein, containing 8 domains, that is anchored in the membrane of the endoplasmic reticulum.<ref name="Roitelman">PMID:1374417</ref> It is the major target of the Statins, a cholesterol lowering drug class and the best selling pharmaceutical drugs in the world. See also [[Ephrin Type-A Receptor]] and [[Neurodevelopmental Disorders]]. | [[HMG-CoA Reductase]] (or '''3-hydroxy-3-methyl-glutaryl-CoA reductase''' or '''HMGR''') is the rate-controlling enzyme of the mevalonate pathway, responsible for cholesterol and other isoprenoid biosynthesis. HMGR is a transmembrane protein, containing 8 domains, that is anchored in the membrane of the endoplasmic reticulum.<ref name="Roitelman">PMID:1374417</ref> It is the major target of the Statins, a cholesterol lowering drug class and the best selling pharmaceutical drugs in the world. See also [[Ephrin Type-A Receptor]] and [[Neurodevelopmental Disorders]]. | ||
| Line 35: | Line 14: | ||
==Structure== | ==Structure== | ||
| - | + | ||
===General Structure=== | ===General Structure=== | ||
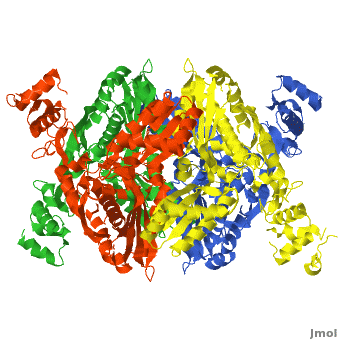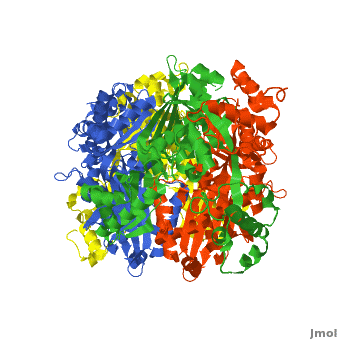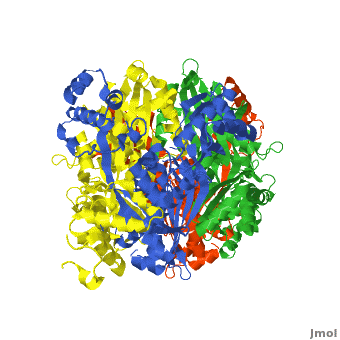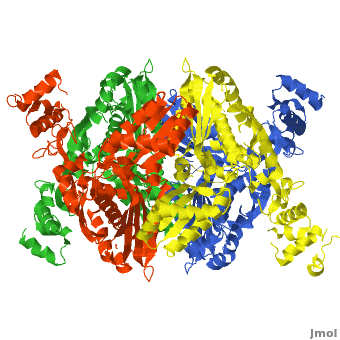
There are two distinct classes of HMGRs, class I HMGRs, which are only found in eukaryotes and are membrane bound and class II HMGRs, which are found in prokaryotes and are soluble.<ref>PMID:11349148</ref> HMGR contains 8 transmembrane domains that have yet to be successfully crystallized, which anchor the protein to the membrane of the endoplasmic reticulum.<ref name="Roitelman"/> The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other.<ref name="Roitelman">PMID:1374417</ref> Within the tetramer, the monomers are arranged into <scene name='HMG-CoA_Reductase/1dq8_2_dimers/3'>two dimers</scene>, each of which contains <scene name='HMG-CoA_Reductase/1dq8_2_active_sites/2'>two active sites </scene>which are formed by residues form both monomers. Each monomer contains <scene name='HMG-CoA_Reductase/1dq8_star3_domains/2'>three domains </scene>, the <scene name='HMG-CoA_Reductase/1dq8_n_domain/2'>N-domain</scene>, the <scene name='HMG-CoA_Reductase/1dq8_l_domain/1'>L-Domain</scene>, and the <scene name='HMG-CoA_Reductase/1dq8_s_domain/1'>S-Domain</scene>. The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of [[ferredoxin]]. The S and L domains are connected by a <scene name='HMG-CoA_Reductase/1dq8_cis_loop/5'>“cis-loop”</scene> which is essential for the HMG-binding site.<ref name="Roitelman"/> Salt bridges between residues R641 and E782 as well as <scene name='HMG-CoA_Reductase/1dq8_cis_loop/4'>hydrogen bonds</scene> between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface.<ref name="Roitelman"/> | There are two distinct classes of HMGRs, class I HMGRs, which are only found in eukaryotes and are membrane bound and class II HMGRs, which are found in prokaryotes and are soluble.<ref>PMID:11349148</ref> HMGR contains 8 transmembrane domains that have yet to be successfully crystallized, which anchor the protein to the membrane of the endoplasmic reticulum.<ref name="Roitelman"/> The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other.<ref name="Roitelman">PMID:1374417</ref> Within the tetramer, the monomers are arranged into <scene name='HMG-CoA_Reductase/1dq8_2_dimers/3'>two dimers</scene>, each of which contains <scene name='HMG-CoA_Reductase/1dq8_2_active_sites/2'>two active sites </scene>which are formed by residues form both monomers. Each monomer contains <scene name='HMG-CoA_Reductase/1dq8_star3_domains/2'>three domains </scene>, the <scene name='HMG-CoA_Reductase/1dq8_n_domain/2'>N-domain</scene>, the <scene name='HMG-CoA_Reductase/1dq8_l_domain/1'>L-Domain</scene>, and the <scene name='HMG-CoA_Reductase/1dq8_s_domain/1'>S-Domain</scene>. The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of [[ferredoxin]]. The S and L domains are connected by a <scene name='HMG-CoA_Reductase/1dq8_cis_loop/5'>“cis-loop”</scene> which is essential for the HMG-binding site.<ref name="Roitelman"/> Salt bridges between residues R641 and E782 as well as <scene name='HMG-CoA_Reductase/1dq8_cis_loop/4'>hydrogen bonds</scene> between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface.<ref name="Roitelman"/> | ||
Revision as of 10:10, 8 May 2013
This page, as it appeared on December 23, 2010, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
Contents |
Medical Implications
| |||||||||||
3D Structures of HMG-CoA Reductase
Updated on 08-May-2013
HGMCR
1r7i - PmHMGCR catalytic domain - Pseudomonas mevalonii
3qae, 3qau - PmHMGCR (mutant) - Streptococcus pneumoniae
HGMCR+statins
3cct, 3ccw, 3ccz, 3cd0, 3cd5, 3cd7, 3cda, 3cdb, 2r4f, 3bgl, 2q1l, 2q6b, 2q6c, 1hw8, 1hw9, 1hwi, 1hwj, 1hwk, 1hwl – HMG-CoA Reductase Catalytic Domain + Statins
1t02 - PmHMGCR catalytic domain + statin derivatives
HGMCR+cofactors
1r31 – PmHMGCR catalytic domain +CoA+mevalonate
1qax - PmHMGCR catalytic domain +HMG+CoA+NAD+
1qay - PmHMGCR catalytic domain +mevalonate+NAD+
1dq8 - hHMGCR catalytic domain (mutant) +CoA+HMG
1dq9 - hHMGCR catalytic domain (mutant)+HMG-CoA
1dqa - hHMGCR catalytic domain (mutant)+HMG+CoA+NADP+
Additional Resources
- See: Pharmaceutical Drug Targets For Additional Information about Drug Targets for Related Diseases
- See: Metabolic Disorders For Additional Information.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Roitelman J, Olender EH, Bar-Nun S, Dunn WA Jr, Simoni RD. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol. 1992 Jun;117(5):959-73. PMID:1374417
- ↑ http://nobelprize.org/nobel_prizes/medicine/laureates/1985/
- ↑ 3.0 3.1 3.2 Meigs TE, Roseman DS, Simoni RD. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J Biol Chem. 1996 Apr 5;271(14):7916-22. PMID:8626470
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005 Sep 16;19(6):829-40. PMID:16168377 doi:10.1016/j.molcel.2005.08.009
- ↑ Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425-30. PMID:1967820 doi:http://dx.doi.org/10.1038/343425a0
- ↑ www.nhlbi.nih.gov/health/.../Diseases/.../CAD_WhatIs.html
- ↑ Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976 Dec 31;72(2):323-6. PMID:16386050
- ↑ http://www.drugs.com/top200.html
- ↑ http://www.medicalnewstoday.com/articles/25046.php
- ↑ Zhang QY, Wan J, Xu X, Yang GF, Ren YL, Liu JJ, Wang H, Guo Y. Structure-based rational quest for potential novel inhibitors of human HMG-CoA reductase by combining CoMFA 3D QSAR modeling and virtual screening. J Comb Chem. 2007 Jan-Feb;9(1):131-8. PMID:17206841 doi:10.1021/cc060101e
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ Corsini A, Maggi FM, Catapano AL. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol Res. 1995 Jan;31(1):9-27. PMID:7784310
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Eran Hodis, Angel Herraez, Joel L. Sussman