We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ramachandran Plot
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
{{clear}} | {{clear}} | ||
== Plot regions limited by steric hindrance == | == Plot regions limited by steric hindrance == | ||
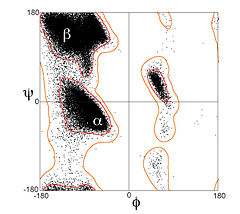
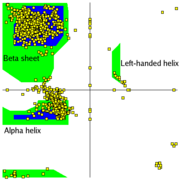
| - | + | Most combinations of φ and ψ are sterically forbidden, <scene name='Ramachandran_Plots/Tripeptide_disallowed/1'>as illustrated in the tripeptide</scene>, '''<font color=limegreen>Glu</font>-Ser-<font color=blue>Ala</font>'''. With Ser having values of φ = 55<sup>o</sup> and ψ = -116<sup>o</sup> the Ser side chain is in contact with the Ala, colored blue. In plots of native peptides the data points will form clusters in the several areas in which steric hindrance does not occur. These regions are illustrated in the Figure on the left, according to our understanding in the early 1990's when the first validation programs such as ProCheck<ref>Laskowski,RA, MacArthur,MW, Moss,DS and Thornton,JM (1993) PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Cryst., 26, 283–291</ref> were developed [[Image:Ramaplot.png|thumb|left|Figure: Ramachandran Plot showing two core regions (blue) and three allowed regions (green).]] [[Image:Ramachandran_general-case_data_and_contours_T8000_small.jpg|thumb|left|240px| General-case Ramachandran plot for >1,000,000 high-quality datapoints]] The core regions (blue in the Figure) contain the most favorable combinations of φ and ψ and contain the greatest number of points. Plots of some proteins contain a small third core region in the upper right quadrant. The allowed regions (green in the Figure) can be located around the core regions or can be unassociated with a core region, but they contain fewer data points than the core regions. The generous regions (not shown in the Figure) extend beyond the allowed regions. The remaining areas are considered disallowed. Observe that the data point 55<sup>o</sup>, -116<sup>o</sup> for Ser of the above tripeptide would fall in the lower right quadrant which contains only a disallowed region in ProCheck. If the Ser in the tripeptide has φ and ψ values of -57<sup>o</sup> and -47<sup>o</sup> which are in the core region in the lower left, the <scene name='Ramachandran_Plots/Tripeptide_allowed/1'>Ser side chain</scene> is rotated away from the Ala and is not in contact with the Ala. Most, if not all, of the points in the above plots for α-helix and β-sheet are in one of the core areas.<p> | |
| - | + | ||
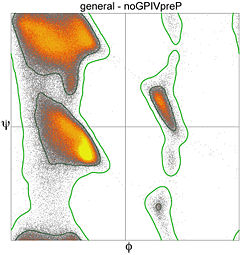
Since the 1990's, great expansion in the number and quality of macromolecular crystal structures and advances in methodology have greatly improved understanding of the energetically favored, allowed, and truly disallowed conformations of proteins and nucleic acids. The lower figure plots Ramachandran values for over a million general-case residues with resolution <2.0Å and backbone B-factor <30. Over half the plot is entirely empty, but there are further clusters evident that have moderate population and presumably have somewhat unfavorable energy but are possible; this actually includes the region near +55, -116, which is found in one type of beta turn. | Since the 1990's, great expansion in the number and quality of macromolecular crystal structures and advances in methodology have greatly improved understanding of the energetically favored, allowed, and truly disallowed conformations of proteins and nucleic acids. The lower figure plots Ramachandran values for over a million general-case residues with resolution <2.0Å and backbone B-factor <30. Over half the plot is entirely empty, but there are further clusters evident that have moderate population and presumably have somewhat unfavorable energy but are possible; this actually includes the region near +55, -116, which is found in one type of beta turn. | ||
Revision as of 08:57, 8 April 2013
This page, as it appeared on November 30, 2010, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
Interactive Ramachandran plots can be generated for any entry in Proteopedia with the use of a typed Jmol command[7]:
- For example, in a new browser window open the entry in Proteopedia for 1bhl
- In the Jmol applet showing the 3D structure on the page, click on the Jmol logo (or frank) in the bottom right corner.
- When the menu comes up, select
Console - Click in the lower text window of the console that comes up and type the command
Ramachandran, followed by the return key. - After some processing the Ramachandran plot will be visible and you can hover over and click on the points in the plot just as you can with atoms in a Jmol scene window. (To return to the model, an easy solution is to reload the page or open a new browser instance of that page, or enter into the console 'model 1.1' without the quotes.) With the console window open, the values will be listed as you click on the spheres.
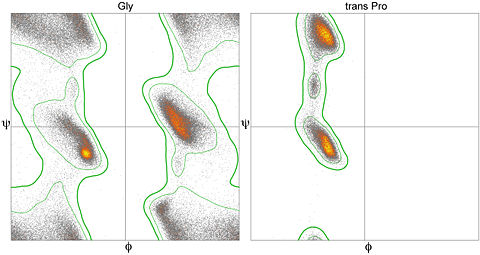
- To limit the plot to displaying certain residues or portions of the structure, you can issue commands in the console, such as 'display helix' or 'display gly'. The latter command will limit the plotted display to just glycine residues. In order to return to showing all values on the plot, issue the command 'display all' in the Jmol console.
If you just need to report φ and ψ values for a few residues, use the Scene Authoring Tools to select the residues of interest and enter the command
draw RAMACHANDRAN in the console.
Notes
- ↑ RAMACHANDRAN GN, RAMAKRISHNAN C, SASISEKHARAN V (July 1963). "Stereochemistry of polypeptide chain configurations". J. Mol. Biol. 7: 95–9. PMID 13990617
- ↑ Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003 Feb 15;50(3):437-50. PMID:12557186 doi:10.1002/prot.10286
- ↑ Laskowski,RA, MacArthur,MW, Moss,DS and Thornton,JM (1993) PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Cryst., 26, 283–291
- ↑ Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003 Feb 15;50(3):437-50. PMID:12557186 doi:10.1002/prot.10286
- ↑ Read RJ, Adams PD, Arendall WB 3rd, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011 Oct 12;19(10):1395-412. PMID:22000512 doi:10.1016/j.str.2011.08.006
- ↑ Ting D, Wang G, Shapovalov M, Mitra R, Jordan MI, Dunbrack RL Jr. Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical Dirichlet process model. PLoS Comput Biol. 2010 Apr 29;6(4):e1000763. PMID:20442867 doi:10.1371/journal.pcbi.1000763
- ↑ Command defined at site for official Jmol documentation
External Resources
- Another example of a Ramachandran Plot showing the different regions. at the European Bioinformatics Institute (EBI)
Proteopedia Page Contributors and Editors (what is this?)
Karl Oberholser, Wayne Decatur, Eran Hodis, Jane S. Richardson, Jaime Prilusky, Alexander Berchansky, Angel Herraez, Norbert Sträter, Joel L. Sussman, Shelly Livne, Eric Martz