We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Michael Roberts/BIOL115 Myo
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
'''SECONDARY STRUCTURE''': | '''SECONDARY STRUCTURE''': | ||
| + | |||
This next view simplifies things, and just shows a <scene name='User:Michael_Roberts/BIOL115_Myo/Secondary_structure/1'>cartoon representation </scene>of the secondary structure of the protein. | This next view simplifies things, and just shows a <scene name='User:Michael_Roberts/BIOL115_Myo/Secondary_structure/1'>cartoon representation </scene>of the secondary structure of the protein. | ||
| Line 26: | Line 27: | ||
'''HYDROPHOBICITY''': | '''HYDROPHOBICITY''': | ||
| + | |||
Globular folds like this are characterised by a polar, <scene name='User:Michael_Roberts/BIOL115_Myo/Secondary_structure/4'>hydrophilic exterior</scene>, which interacts with the aqueous solvent, and a hydrophobic core. | Globular folds like this are characterised by a polar, <scene name='User:Michael_Roberts/BIOL115_Myo/Secondary_structure/4'>hydrophilic exterior</scene>, which interacts with the aqueous solvent, and a hydrophobic core. | ||
| + | {{Template:ColorKey_Hydrophobic}}, {{Template:ColorKey_Polar}} | ||
| - | + | The next view shows a section through the protein that highlights the <scene name='User:Michael_Roberts/BIOL115_Myo/Secondary_structure/5'>hydrophobic core </scene>better. | |
This view has been produced in the software by a process known as 'slabbing'. You can still rotate the molecule around - whatever view you see will the the front part of the view of the protein cut off. | This view has been produced in the software by a process known as 'slabbing'. You can still rotate the molecule around - whatever view you see will the the front part of the view of the protein cut off. | ||
'''THE HEME GROUP''': | '''THE HEME GROUP''': | ||
| + | |||
Now let's turn our attention to the main function of myoglobin - oxygen binding. | Now let's turn our attention to the main function of myoglobin - oxygen binding. | ||
Oxygen is bound by a <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/1'>heme group</scene>, which sits in a hydrophobic pocket in the myoglobin protein. | Oxygen is bound by a <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/1'>heme group</scene>, which sits in a hydrophobic pocket in the myoglobin protein. | ||
| + | |||
Central to the heme group is an <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/3'>iron (Fe) atom</scene>. | Central to the heme group is an <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/3'>iron (Fe) atom</scene>. | ||
| + | |||
'''PROXIMAL AND DISTAL HISTIDINES''': | '''PROXIMAL AND DISTAL HISTIDINES''': | ||
| + | |||
The iron atom sits either side of the side chains of two <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/4'>histidine residues</scene>. | The iron atom sits either side of the side chains of two <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/4'>histidine residues</scene>. | ||
| + | One of these (coloured cyan) is attached to the iron atom, and is known as the ''proximal'' histidine. The other (green) is called the ''distal'' histidine. | ||
| + | |||
| + | |||
| + | '''OXYGEN''': | ||
| + | The space between the iron and the distal histidine is where the <scene name='User:Michael_Roberts/BIOL115_Myo/Heme/6'>oxygen </scene>binds. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 16:09, 12 April 2013
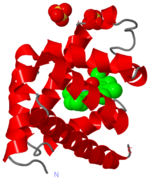
Crystal Structure of myoglobin, 1a6m
The heme group and oxygen binding in myoglobin.
Myoglobin is a protein whose function is to store oxygen in muscle tissues. Like heamoglobin, it is red in colour, and it is myoglobin that gives muscle its strong red colour.
Myoglobin was the first globular protein for which the 3-dimensional structure was solved, back in the late 1950s. It gives its name to the 'globin fold', a common alpha domain motif. An alpha domain is a structural region composed entirley of alpha-helix.
Click on the 'green links' in the text in the scrollable section below to examine this molecule in more detail.
| |||||||||||
