HMG-CoA Reductase
From Proteopedia
| Line 22: | Line 22: | ||
The HMG-CoA and NADPH molecules make numerous contacts with the <scene name='HMG-CoA_Reductase/Ls_domains/3'>L and S domains</scene> in forming the four active sites. The CoA is located in a <scene name='HMG-CoA_Reductase/1dqa_nadp_and_coa/2'>near the enzyme surface</scene>, with the pantothenic acid moiety extending into the interior of the protein. <scene name='HMG-CoA_Reductase/1dqa_tyr_491/2'>Tyrosine 479 forms a hydrophobic lid</scene> over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with <scene name='HMG-CoA_Reductase/1dqa_loop/2'>a loop region</scene> playing a critical role in binding.<ref name="Roitelman"/> | The HMG-CoA and NADPH molecules make numerous contacts with the <scene name='HMG-CoA_Reductase/Ls_domains/3'>L and S domains</scene> in forming the four active sites. The CoA is located in a <scene name='HMG-CoA_Reductase/1dqa_nadp_and_coa/2'>near the enzyme surface</scene>, with the pantothenic acid moiety extending into the interior of the protein. <scene name='HMG-CoA_Reductase/1dqa_tyr_491/2'>Tyrosine 479 forms a hydrophobic lid</scene> over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with <scene name='HMG-CoA_Reductase/1dqa_loop/2'>a loop region</scene> playing a critical role in binding.<ref name="Roitelman"/> | ||
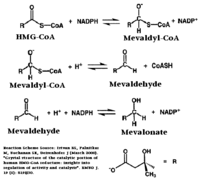
| - | The HMG binding pocket is the site of catalysis in HMGR. <scene name='HMG-CoA_Reductase/1dqa_cis_loop2/2'> | + | The HMG binding pocket is the site of catalysis in HMGR. <scene name='HMG-CoA_Reductase/1dqa_cis_loop2/2'> The “cis-loop” that bends over the top of HMG </scene> ([[1dqa]]) is a critical structural element of this binding site. Residues <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> and are positioned in the active site as is <scene name='HMG-CoA_Reductase/1dqa_k691/2'>K691 which is only 2.7 angstroms from the HMG O2 carbonyl oxygen</scene>. It is this K691 that likely stabilizes the negatively charged oxygen of the first mevaldyl-CoA intermediate.<ref name="Roitelman"/> The mevaldyl CoA intermediate is subsequently converted to Mavaldehyde with added stabilization from <scene name='HMG-CoA_Reductase/1dqa_h866/2'>H866, which is within hydrogen bonding distance of the thiol group</scene>. It is then believed that the close proximity of <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> increases the pKA of E559, allowing it to be a proton donor for the reduction of mevaldehyde into mevalonate.<ref name="Roitelman"/> |
<br /> | <br /> | ||
| Line 36: | Line 36: | ||
==Medical Implications== | ==Medical Implications== | ||
| - | + | ||
===Medical Implications for HMGR=== | ===Medical Implications for HMGR=== | ||
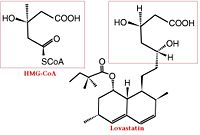
[[Image: HMGCoA to Lovastatin Comparison.jpg|200px|left|thumb| Comparison of Chemical Structure of HMG-CoA and Lovastatin]] | [[Image: HMGCoA to Lovastatin Comparison.jpg|200px|left|thumb| Comparison of Chemical Structure of HMG-CoA and Lovastatin]] | ||
Revision as of 10:12, 8 May 2013
This page, as it appeared on December 23, 2010, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
Contents |
Medical Implications
Medical Implications for HMGR
Elevated cholesterol levels have been identified as a major risk factor for coronary artery disease, the narrowing of arteries of the heart, which affected over 13 million people in the United States alone. It is a major cause of disability and death, killing over 500 thousand people in the USA in 2001.[7]
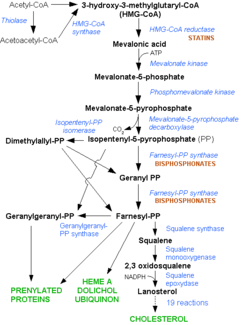
The statins are HMG-CoA reductase inhibitors. Discovered by Akira Endo in 1971, statins are similar in structure to HMG-CoA and act by competitively inhibiting HMGR. Since HMGR is the first committed enzyme in the cascade that eventually produces cholesterol, use of statins can dramatically reduce blood cholesterol levels.[8] As a drug class, statins generated over $20 billion in sales in 2009 with Pfizer’s Lipitor being the best selling drug in the world at the time of writing.[9][10] Statin development stands as a triumph for rational drug design validating rational drug design as a proven approach toward creating selective, therapeutic compounds.[11]
A number of crystal structures of HMGR with bound statins have been solved which elucidate how the statin molecule is bound by HMGR. Statins in general occupy the active site of HMGR, preventing HMG-CoA from binding. The structure of HMGR with bound statins ( (1hwk) marketed as Lipitor) shows that the cis loop forms a number of polar interactions with the statin inhibitor, particularly residues Ser 684, Asp 690, Lys 691, Lys 692, and hydrogen bond interactions between Glu 559 and Asp 767 with the O5-hydroxyl of the statins. Van der Waals interactions between Leu 562, Val 683, Leu 853, Ala 856, and Leu 857 of HMGR and hydrophobic ring structures of the statins contribute to binding as well.[12] These interactions result in the statins binding to HMGR with a Ki of between .1-2.3nM while the Michaelis constant KM for HMG-CoA is 4uM, allowing the statins to outcompete HMG-CoA in binding to HMGR.[13] Additional structures of HMGR with the statins (1hw8), (1hw9, Zocor), (1hwi, Lescol), (1hwj, Baycol), and (1hwl, Crestor) highlight the important residues involved in inhibitor binding. Other statins approved by the FDA include Lovastatin.
</StructureSection>
3D Structures of HMG-CoA Reductase
Updated on 08-May-2013
HGMCR
1r7i - PmHMGCR catalytic domain - Pseudomonas mevalonii
3qae, 3qau - PmHMGCR (mutant) - Streptococcus pneumoniae
HGMCR+statins
3cct, 3ccw, 3ccz, 3cd0, 3cd5, 3cd7, 3cda, 3cdb, 2r4f, 3bgl, 2q1l, 2q6b, 2q6c, 1hw8, 1hw9, 1hwi, 1hwj, 1hwk, 1hwl – HMG-CoA Reductase Catalytic Domain + Statins
1t02 - PmHMGCR catalytic domain + statin derivatives
HGMCR+cofactors
1r31 – PmHMGCR catalytic domain +CoA+mevalonate
1qax - PmHMGCR catalytic domain +HMG+CoA+NAD+
1qay - PmHMGCR catalytic domain +mevalonate+NAD+
1dq8 - hHMGCR catalytic domain (mutant) +CoA+HMG
1dq9 - hHMGCR catalytic domain (mutant)+HMG-CoA
1dqa - hHMGCR catalytic domain (mutant)+HMG+CoA+NADP+
Additional Resources
- See: Pharmaceutical Drug Targets For Additional Information about Drug Targets for Related Diseases
- See: Metabolic Disorders For Additional Information.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Roitelman J, Olender EH, Bar-Nun S, Dunn WA Jr, Simoni RD. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol. 1992 Jun;117(5):959-73. PMID:1374417
- ↑ http://nobelprize.org/nobel_prizes/medicine/laureates/1985/
- ↑ 3.0 3.1 3.2 Meigs TE, Roseman DS, Simoni RD. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J Biol Chem. 1996 Apr 5;271(14):7916-22. PMID:8626470
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005 Sep 16;19(6):829-40. PMID:16168377 doi:10.1016/j.molcel.2005.08.009
- ↑ Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425-30. PMID:1967820 doi:http://dx.doi.org/10.1038/343425a0
- ↑ www.nhlbi.nih.gov/health/.../Diseases/.../CAD_WhatIs.html
- ↑ Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976 Dec 31;72(2):323-6. PMID:16386050
- ↑ http://www.drugs.com/top200.html
- ↑ http://www.medicalnewstoday.com/articles/25046.php
- ↑ Zhang QY, Wan J, Xu X, Yang GF, Ren YL, Liu JJ, Wang H, Guo Y. Structure-based rational quest for potential novel inhibitors of human HMG-CoA reductase by combining CoMFA 3D QSAR modeling and virtual screening. J Comb Chem. 2007 Jan-Feb;9(1):131-8. PMID:17206841 doi:10.1021/cc060101e
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ Corsini A, Maggi FM, Catapano AL. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol Res. 1995 Jan;31(1):9-27. PMID:7784310
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Eran Hodis, Angel Herraez, Joel L. Sussman