FK506 binding protein
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='3jym.pdb' size='450' frame='true' align='right' scene='3jym/Cv/2' caption='Wheat FKBP506 [[3jym]]'> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
'''FK506 binding protein (FKBP)''' is a prolyl isomerase related to the [[Cyclophilin|cyclophilins]]. FKBP is a folding chaperone for proteins containing prolines. FKBP12 binds the immunosuppressor tacrolimus (ascomycin FK506) which is used against organ rejection and rapamycin. For more details see<br /> | '''FK506 binding protein (FKBP)''' is a prolyl isomerase related to the [[Cyclophilin|cyclophilins]]. FKBP is a folding chaperone for proteins containing prolines. FKBP12 binds the immunosuppressor tacrolimus (ascomycin FK506) which is used against organ rejection and rapamycin. For more details see<br /> | ||
* [[Human FKBP52]]<br /> | * [[Human FKBP52]]<br /> | ||
| Line 31: | Line 5: | ||
* [[Wheat FKBP73]]. | * [[Wheat FKBP73]]. | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
| - | <br /> | ||
==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ||
| - | |||
| - | <StructureSection load='3jym.pdb' size='350' frame='true' align='right' scene='3jym/Cv/2' caption='Wheat FKBP506 [[3jym]]'> | ||
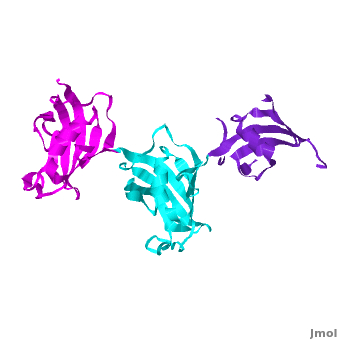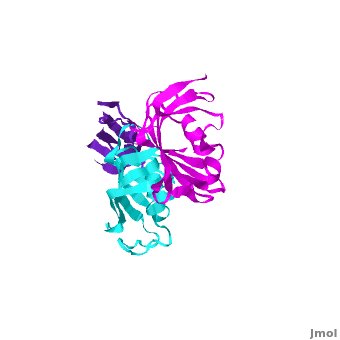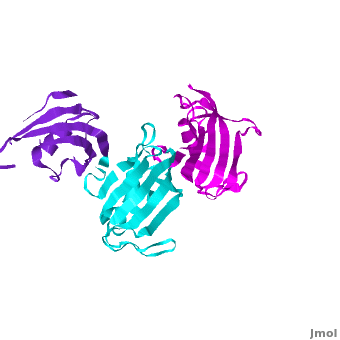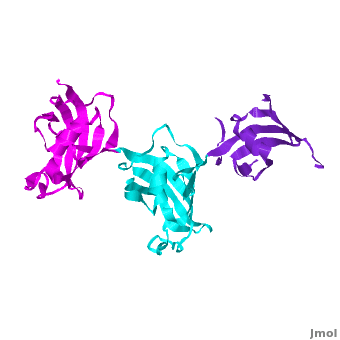
Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, <font color='cyan'><b>wFK73_2 (residues 149–266) in cyan</b></font> and <font color='magenta'><b>wFK73_3 (residues 267–386) in magenta</b></font> ([[3jym]]). The wFK73_1 domain exhibits electron density only between residues 33–38, 54–69 and 87–148. The bulges and the flaps as well as the N- and C-termini are labeled. The three FK506 binding (FK) domains of wFKBP73 are held together mainly by <scene name='3jym/Cv/4'>salt bridge networks</scene> situated between each pair of domains. The wFK73_2-wFK73_1 domains are held by a salt bridge between Lys162–Glu62, and a salt bridge network between Arg151–Asp61 and Glu58. The interface between wFK73_2-wFK73_3 is held by two salt bridges between Lys204–Glu269, and Glu178–Lys279. The interactions Lys162–Glu62 and Glu178–Lys279, involve conserved residues (Glu62 from wFK73_1 and Glu178 from wFK73_2, Lys162 from wFK73_2 and Lys279 from wFK73_3). | Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, <font color='cyan'><b>wFK73_2 (residues 149–266) in cyan</b></font> and <font color='magenta'><b>wFK73_3 (residues 267–386) in magenta</b></font> ([[3jym]]). The wFK73_1 domain exhibits electron density only between residues 33–38, 54–69 and 87–148. The bulges and the flaps as well as the N- and C-termini are labeled. The three FK506 binding (FK) domains of wFKBP73 are held together mainly by <scene name='3jym/Cv/4'>salt bridge networks</scene> situated between each pair of domains. The wFK73_2-wFK73_1 domains are held by a salt bridge between Lys162–Glu62, and a salt bridge network between Arg151–Asp61 and Glu58. The interface between wFK73_2-wFK73_3 is held by two salt bridges between Lys204–Glu269, and Glu178–Lys279. The interactions Lys162–Glu62 and Glu178–Lys279, involve conserved residues (Glu62 from wFK73_1 and Glu178 from wFK73_2, Lys162 from wFK73_2 and Lys279 from wFK73_3). | ||
| Line 53: | Line 14: | ||
It was shown that 12 conserved residues (for wFK73_1 domain they are Tyr67, Phe77, Asp78, Arg83, Phe87, Gln95, Val96, Ile97, Trp100, Tyr123, Ile132, and Phe140) of the FK1 domains of hFKBP12, 13, 25, 51 and 52, are involved in binding the FK506 or rapamycin. Since only the FK1 domains contain all the conserved amino acids (in contrast to FK2 and/or FK3 domains), only they exhibit PPIase activity, which can be inhibited by the binding of the drugs FK506, and rapamycin. These conserved residues form the hydrophobic cavity. The structure of hFKBP12 ([[2ppn]]) demonstrates a good example of this <scene name='3jym/Cavity/1'>cavity</scene>. All these residues are conserved in the wFK73_1 domain, it could be assumed that a similar cavity is also formed in wFK73_1, although some of these residues are missing electron density in the wFK73 structure and, therefore, it can not be seen. Domain <font color='magenta'><b>wFK73_3</b></font> has <scene name='3jym/Cavity/3'>narrower cavity</scene>, whereas <font color='cyan'><b>wFK73_2</b></font> <scene name='3jym/Cavity/4'>lacks this cavity at all</scene>. Conserved residues are colored yellow. So, the lack of drug binding of the wFK73_2 and wFK73_3 domains could be explained by the absence of the conserved drug binding residues. This is in agreement with the fact that the FK2 domains of hFKBP51 and hFKBP52 and the single FK domains of FKBP38, DmFKBP45 and AtFKBP42, all lacking the conserved residues, do not exhibit drug binding. | It was shown that 12 conserved residues (for wFK73_1 domain they are Tyr67, Phe77, Asp78, Arg83, Phe87, Gln95, Val96, Ile97, Trp100, Tyr123, Ile132, and Phe140) of the FK1 domains of hFKBP12, 13, 25, 51 and 52, are involved in binding the FK506 or rapamycin. Since only the FK1 domains contain all the conserved amino acids (in contrast to FK2 and/or FK3 domains), only they exhibit PPIase activity, which can be inhibited by the binding of the drugs FK506, and rapamycin. These conserved residues form the hydrophobic cavity. The structure of hFKBP12 ([[2ppn]]) demonstrates a good example of this <scene name='3jym/Cavity/1'>cavity</scene>. All these residues are conserved in the wFK73_1 domain, it could be assumed that a similar cavity is also formed in wFK73_1, although some of these residues are missing electron density in the wFK73 structure and, therefore, it can not be seen. Domain <font color='magenta'><b>wFK73_3</b></font> has <scene name='3jym/Cavity/3'>narrower cavity</scene>, whereas <font color='cyan'><b>wFK73_2</b></font> <scene name='3jym/Cavity/4'>lacks this cavity at all</scene>. Conserved residues are colored yellow. So, the lack of drug binding of the wFK73_2 and wFK73_3 domains could be explained by the absence of the conserved drug binding residues. This is in agreement with the fact that the FK2 domains of hFKBP51 and hFKBP52 and the single FK domains of FKBP38, DmFKBP45 and AtFKBP42, all lacking the conserved residues, do not exhibit drug binding. | ||
| - | See also [[Wheat FKBP73]]. | ||
| - | |||
| - | </StructureSection> | ||
==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ||
| - | <StructureSection load='2kr7' size='350' side='right' scene='Journal:JBIC:14/Cv/2' caption='SlyD [[2kr7]]'> | ||
SlyD belongs to the FK506-binding protein (FKBP) family with both peptidylprolyl isomerase (PPIase) and chaperone activities, and is considered to be a ubiquitous cytosolic protein-folding facilitator in bacteria. It possesses a histidine- and cysteine-rich C-terminus binding to selected divalent metal ions (''e.g.'', Ni<sup>2+</sup>, Zn<sup>2+</sup>), which is important for its involvement in the maturation processes of metalloenzymes. The solution structure of <scene name='Journal:JBIC:14/Cv/3'>C-terminus-truncated SlyD</scene> from ''Helicobacter pylori'' (HpSlyDΔC) was determined ([[2kr7]]). HpSlyDΔC folds into <scene name='Journal:JBIC:14/Cv/4'>two well-separated, orientation-independent domains:</scene> the <span style="color:cyan;background-color:black;font-weight:bold;">PPIase-active FKBP domain (in cyan)</span> and the <font color='red'><b>chaperone-active insert-in-flap (IF) domain (in red)</b></font>, <font color='darkmagenta'><b>linkers are in darkmagenta</b></font>. The FKBP domain consists of a four-stranded antiparallel <scene name='Journal:JBIC:14/Cv/5'>β-sheet with an α-helix on one side, whereas the IF domain folds into a four-stranded antiparallel β-sheet accompanied by a short α-helix.</scene> Intact ''H. pylori'' SlyD binds both Ni<sup>2+</sup> and Zn<sup>2+</sup>, with dissociation constants of 2.74 and 3.79 μM respectively. Intriguingly, binding of Ni<sup>2+</sup> instead of Zn<sup>2+</sup> induces protein conformational changes around the <scene name='Journal:JBIC:14/Cv/6'>active sites of the FKBP domain, implicating a regulatory role of nickel</scene> <font color='blueviolet'><b>(residues experiencing relatively large chemical shift perturbations upon interactions of HpSlyDΔC with Ni<sup>2+</sup> are in blueviolet)</b></font>. <scene name='Journal:JBIC:14/Cv/7'>The twin-arginine translocation (Tat) signal peptide from the small subunit of [NiFe] hydrogenase (HydA) binds the protein at the IF domain</scene> <font color='orange'><b>(residues in orange)</b></font>. Surprisingly, several residues (Ile41, Gly42, Ile46, and Asn31) were from the FKBP domain, which is likely due to the binding of the longer n-region of HydA Tat peptide to the FKBP domain. Nickel binding and the recognition of the Tat signal peptide by the protein suggest that SlyD participates in [NiFe] hydrogenase maturation processes. | SlyD belongs to the FK506-binding protein (FKBP) family with both peptidylprolyl isomerase (PPIase) and chaperone activities, and is considered to be a ubiquitous cytosolic protein-folding facilitator in bacteria. It possesses a histidine- and cysteine-rich C-terminus binding to selected divalent metal ions (''e.g.'', Ni<sup>2+</sup>, Zn<sup>2+</sup>), which is important for its involvement in the maturation processes of metalloenzymes. The solution structure of <scene name='Journal:JBIC:14/Cv/3'>C-terminus-truncated SlyD</scene> from ''Helicobacter pylori'' (HpSlyDΔC) was determined ([[2kr7]]). HpSlyDΔC folds into <scene name='Journal:JBIC:14/Cv/4'>two well-separated, orientation-independent domains:</scene> the <span style="color:cyan;background-color:black;font-weight:bold;">PPIase-active FKBP domain (in cyan)</span> and the <font color='red'><b>chaperone-active insert-in-flap (IF) domain (in red)</b></font>, <font color='darkmagenta'><b>linkers are in darkmagenta</b></font>. The FKBP domain consists of a four-stranded antiparallel <scene name='Journal:JBIC:14/Cv/5'>β-sheet with an α-helix on one side, whereas the IF domain folds into a four-stranded antiparallel β-sheet accompanied by a short α-helix.</scene> Intact ''H. pylori'' SlyD binds both Ni<sup>2+</sup> and Zn<sup>2+</sup>, with dissociation constants of 2.74 and 3.79 μM respectively. Intriguingly, binding of Ni<sup>2+</sup> instead of Zn<sup>2+</sup> induces protein conformational changes around the <scene name='Journal:JBIC:14/Cv/6'>active sites of the FKBP domain, implicating a regulatory role of nickel</scene> <font color='blueviolet'><b>(residues experiencing relatively large chemical shift perturbations upon interactions of HpSlyDΔC with Ni<sup>2+</sup> are in blueviolet)</b></font>. <scene name='Journal:JBIC:14/Cv/7'>The twin-arginine translocation (Tat) signal peptide from the small subunit of [NiFe] hydrogenase (HydA) binds the protein at the IF domain</scene> <font color='orange'><b>(residues in orange)</b></font>. Surprisingly, several residues (Ile41, Gly42, Ile46, and Asn31) were from the FKBP domain, which is likely due to the binding of the longer n-region of HydA Tat peptide to the FKBP domain. Nickel binding and the recognition of the Tat signal peptide by the protein suggest that SlyD participates in [NiFe] hydrogenase maturation processes. | ||
Revision as of 10:28, 10 November 2013
| |||||||||||
Contents |
3D Structures of FKBP
Updated on 10-November-2013
FKBP
1fks, 1fkr, 1fkt – EcFKBP – Escherichia coli
1q6h, 1q6u – EcFKBP
1q6i – EcFKBP+FK-506
1ix5 – FKBP – Methanothermococcus thermolithotrophicus – NMR
1bl4 – hFKBP (mutant)+inhibitor
1bkf – hFKBP (mutant)+FK-506
1fkf - hFKBP+FK-506
1fkr, 1fks, 1fkt – hFKBP - NMR
FKBP1
3mdy – hFKBP + BMPR1B
3h9r - hFKBP-1A+ activin receptor type I
1j4r – hFKBP-1+FKB-001
FKBP3
3kz7 – hFKBP FK506-binding domain + immunosuppressant - human
FKBP4
1q1c – hFKBP
1n1a – hFKBP N terminal
1p5q - hFKBP C terminal
1qz2 – hFKBP + Hsp90 peptide
4drj – hFKBP FK506-binding domain + serine/threonine-protein kinase FRB domain + Immunospressant
FKBP5
3o5d, 3o5e, 3o5f – hFKBP
3o5g, 3o5i, 3o5j, 3o5k - hFKBP FK506-binding domain
3o5l, 3o5m, 3o5o, 3o5p, 3o5q - hFKBP FK506-binding domain (mutant)
3o5r - hFKBP FK506-binding domain (mutant) + immunosuppressant
4drq – hFKBP + pipecolate sulfonamide
4drk, 4drm, 4drn, 4dro, 4drp – hFKBP FK506-binding domain + FK506 analog
FKBP8
2f2d, 3ey6, 2awg - hFKBP FK506-binding domain
2d9f – hFKBP – NMR
2jwx - hFKBP N terminal - NMR
FKBP12
1eym, 2ppo, 4ipx – hFKBP (mutant)
1fkk – hFKBP
2gaq, 2pnu– hFKBP - NMR
3uqi – ymFKBP – yellowfever mosquito
2lpv – ymFKBP - NMR
1fkd, 1fkj, 2fke, 1qpf, 1qpl – hFKBP + immunosuppressant
2ppp, 2ppn, 2dg3, 1d6o – hFKBP
1j4h, 1j4i – hFKBP + inhibitor
1b6c – hFKBP + TGF-B superfamily receptor I
3fap – hFKBP + FKBP12-rapamycin associated protein
4fap - hFKBP + FKBP12-rapamycin associated protein + immunosuppressant
1tco - FKBP + Ser/Thr phosphatase B2 + immunosuppressant - bovine
1yat – FKBP + antagonist – yeast
3uqi – ymFKBP – yellowfever mosquito
2lpv – ymFKBP - NMR
FKBP26
3pr9, 3pra, 3prb, 3prd – FKBP – Methanocaldococcus jannaschii
FKBP35
2vn1 – PfFKBP FK506-binding domain+FK506
FKBP36
3b7x – hFKBP
FKBP38
2jwx - hFKBP N terminal - NMR
FKBP42
2f4e – AtFKBP N-terminal
2if4 – AtFKBP
FKBP51
1kt0, 1kt1 – hFKBP
4drq – hFKBP + pipecolate sulfonamide
4drk, 4drm, 4drn, 4dro, 4drp – hFKBP + FK506 analog
4drh, 4dri – hFKBP FK1 domain + serine/threonine protein kinase MTOR peptide + rapamycin
FKBP52
4drj – hFKBP FK1 domain + serine/threonine protein kinase MTOR peptide + rapamycin
FKBP59
1rot, 1rou – FKBP N terminal – NMR – rabbit
2kr7 – FKBP SlyD – NMR - Helicobacter pylori
2lgo – FKBP – NMR –Giardia lamblia
FKBP73
- ↑ Unger T, Dym O, Albeck S, Jacobovitch Y, Bernehim R, Marom D, Pisanty O, Breiman A. Crystal structure of the three FK506 binding protein domains of wheat FKBP73: evidence for a unique wFK73_2 domain. J Struct Funct Genomics. 2010 Jun;11(2):113-23. Epub 2010 Mar 20. PMID:20306145 doi:10.1007/s10969-010-9085-8
- ↑ Cheng T, Li H, Xia W, Sun H. Multifaceted SlyD from Helicobacter pylori: implication in [NiFe] hydrogenase maturation. J Biol Inorg Chem. 2011 Nov 2. PMID:22045417 doi:10.1007/s00775-011-0855-y