A comparison of the Replication Terminator Protein (from Bacillus subtilis) and Tus (from Escherichia coli) provides an interesting insight into how proteins with vastly different structures and mechanisms of action can produce essentially identical effects in their native systems.
Looking at the structures of these two proteins, it is not immediately obvious that they would perfom the same function; to arrest the progression of the replication fork along the bacterial chromosome at specific sites (Ter sites). Furthermore, this arrest-mechanism functions in a polar manner in both organisms, which is perhaps surprising considering the symmetrical characteristics of both proteins.[1]
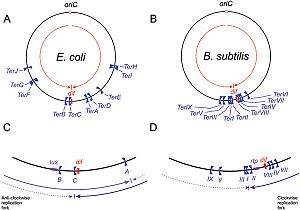
Schematic representation of
Ter sites in
B. subtilis and
E. coli, Taken from Duggin
et al (2008).
The Replication Fork and Polar Arrest
DNA replication of circular bacterial chromosomes occurs using two replication forks that originate from a single location (oriC) and move in opposite directions around the chromosome. In E. coli, B. subtilis, and other bacteria and archaea, these replication forks are halted by interactions with terminator proteins bound to DNA sites known as "Terminator sites", orTer sites. The termination of the replication fork is dependent on the direction of approach to these Ter sites: if the replication fork approaches from the permissive face replication will continue; however, if the replication fork approaches from the non-permissive face the fork will be arrested and DNA replication will cease at that point. While it is possible for these organisms to function without this type of replication-arrest mechanism, the conservation of this system across species indicates some form of evolutionary benefit.
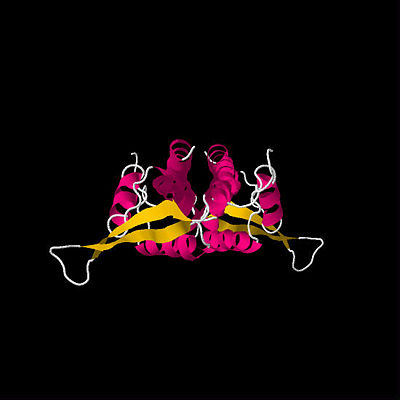
First determined RTP structure. By Bussiere
et al.(1995)

Unlocked Tus-
Ter complex, detemined by Kamada
et al.(1996)
RTP: A homodimer responsible for polar arrest
The Replication Terminator Protein (RTP) from Bacillus subtilis is comprised of two identical monomers 14.5 kDa in size which bind to DNA to form a homodimer. The separate monomers bind at 30 bp sequences known as the A and B termination (Ter) sites. Both of these sites have inverted 16 bp repeats which overlap at highly conserved TAT trinucleotide sequence. The structure of RTP is commonly referred to as a “winged helix” DNA binding motif and consists of a compact α helix / β-strand with a protruding loop (or “wing”) between the β2 and β3 strands. Both monomers of RTP interact with DNA specifically through hydrogen bonding at residues
, present in the α3 recognition helix. RTP also forms non-specific interactions at its N-terminus region.[2]
The first crystal structure of RTP was determined in 1995 by Bussiere et al. (See figure above).[3] This initial structure, which used a symmetric B Ter DNA homologue, suggested that RTP exists as a symmetric homodimer. The idea that a symmetric protein structure could be responsible for an inherently polar mechanism has resulted in a series of proposed solutions and discoveries regarding the mechanism of replication fork arrest.
Induced Conformational Change
Early theories to explain replication fork arrest by RTP involved the concept of a “molecular clamp”, which suggests that the affinity between the Ter DNA and the RTP protein is sufficient to stop the replication fork. One of the earliest theories was the Induced Conformational Change Model. This model, first described by Smith et al. in 1996, proposes that when the first monomer of RTP binds the Ter-B site on the chromosome it causes slight DNA bending, which promotes the binding of the second monomer of RTP to the A site via cooperative binding. According to this model, the binding of the second monomer to the A site changes conformation of Ter-B-RTP complex such that it is able to inhibit DNA unwinding by the helicase enzyme and arrest the replication fork.[4]
Differential Binding Affinity
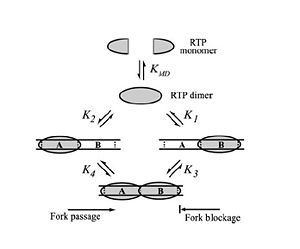
Differential Binding Affinity Model proposed by Kralicek
et al. in 1997. Image from Duggin
et al., 2004.
In 1997, Kralicek et al. proposed an alternate theory of RTP arrest known as the Differential Binding Affinity Model. This model also involves the idea of a “molecular clamp”, and states that the polar arrest mechanism can be explained purely based on the differential binding affinities of RTP to the A and B termination sites. The theory is based on the assumption that the affinity of RTP for the B site in the complete complex is much greater than the affinity of for the A site in the complete complex, or the affinity of a single RTP monomer to the B site alone. According to the model, only the affinity of the complex RTP for the Ter-B (K3 in Figure X) is sufficient to prevent the removal of RTP from DNA when the replication fork moves along the DNA; therefore, if the replisome approaches from the B site, the RTP is not removed from the DNA and the replication fork is arrested. Similarly, when the replication fork approaches from the A site the binding affinity (K4 in Figure X) is not sufficient to prevent the removal of RTP and the replisome is able to pass.[5]
The idea that RTP binds with differing affinity to the A and B Ter sites has since been explained on a molecular level with the determination of the crystal structure of RTP while bound to its native B Ter site by Vivian et al., 2007. This structure differed from that found by Bussiere et al. in that it used RTP bound to the native B Ter site (nRB), which is asymmetric, as opposed to a symmetric homologue (sRB). This revealed both the protein and the Ter DNA are asymmetric, explaining the differential binding affinities between the A and B Ter sites.[6]
The entire concept of a “molecular clamp” in fork arrest has since been challenged by mutational studies performed by Duggin et al. in 2004. After creating mutant DNA Ter sites and analysing the resulting efficiency of replication fork arrest, Duggin et al. found that mutations which caused decreased affinity of RTP for the proximal half of the terminator DNA (i.e. the half which faces the approaching replisome) did not necessarily decrease fork arrest efficiency, and that increased proximal site affinity did not increase fork arrest efficiency. These results were inconsistent with the differential binding affinity model and induced conformational change, suggesting other factors apart from DNA-protein binding must also be responsible for replication fork arrest by RTP.[7]
Is replication fork arrest by RTP affinity independent?
This mutational data provided by Duggin et al. (2004) suggested that replication fork arrest was a more complex process than one based purely on binding between RTP and DNA. In 2009, Duggin proposed the alternate theory that the C-terminus of an RTP monomer is able to contact the oncoming helicase and that this interaction is responsible for replication fork arrest. To test this hypothesis, Duggin created a series of RTP fusion proteins which were constructed by adding a GFP peptide to the C-terminus using a variety of spacer amino acids. Because the C-terminus is not in contact with DNA, the fusion proteins had no affect on the DNA binding properties of RTP; however, the fusion proteins had significantly reduced fork arrest efficiencies. These results effectively rule out the molecular clamp model, showing that RTP activity is independent of DNA binding affinity and suggesting that this C-terminal interaction with helicase is responsible for the fork arrest properties of RTP.[8]
Tus: An asymmetric monomer and unlikely candidate.
In 1996, Kamada et al. determined the crystal structure Tus bound to a 16bp fragment of Ter DNA, revealing that Tus binds Ter DNA as an asymmetrical monomer with no identifiable DNA-binding motifs. Tus consists of three distinct structural regions: two α-helical regions and a central β-strand, which together form a large, positively-charged central cleft. The core β-sheets interact with 13 base pairs of duplex Ter-DNA by partial insertion into the major groove, and at least 30 other residues make nonspecific contacts with the DNA backbone.
The positioning of α-helices in the Tus protein is particularly notable: two protrude from both the amino and carboxy domains to clasp the DNA duplex, thereby shielding the interdomain β structures from direct contacts with other proteins (such as the DnaB helicase).[9] The concentration of α-helices on the non-permissive face of Tus is absolutely cruical to the protein's ability to form a locked complex with the Ter site (as discussed later).
Four models of replication fork arrest
In a review paper in 2005, Neylon et al. summarised the four proposed mechanisms of polar arrest by the Tus-Ter complex:
1. The Clamp model - Tus acts as a simple thermodynamic clamp which blocks DnaB progression.
2. The Interaction model - Tus directly interacts with DnaB to prevent its progression.
3. Tus engineers a DNA structure on the non-permissive face that is a physical block to DnaB
4. DnaB engineers a DNA structure on the permissive face that actively promotes Tus dissociation.
Neylon et al. decided that the Clamp model was too simplistic to explain the polar nature of fork arrest. Based on mutational data, they concluded that it is probably a combination of Tus-DnaB interactions as well as Tus-Ter binding strength that contribute to fork-arrest activity.
A stepwise model of dissociation of Tus from Ter DNA appealed to Neylon et al. This would involve the formation of a nonspecific Tus-DNA complex before the formation of a specific Tus-Ter complex during binding. According to this model, as DnaB approaches from the permissive end it promotes the formation of the lower-affinity nonspecific complex, which would then rapidly dissociate; conversely, when DnaB approaches from the nonpermissive face, formation of the nonspecific complex would be prevented and Tus would become kinetically locked onto the Ter DNA.[10]
So how does Tus stop the replication fork, and why is it a polar arrest mechanism?
Mulcair et al. (2006) discovered that the key to Tus forming a locked complex with Ter was twofold: firstly, the locked complex was formed only on the approach of DnaB helicase (the leading edge of the replication fork); and secondly, this locked complex was due to the base-flipping of C6 of Ter DNA into a cytosine-specific binding pocket on Tus. The approach of DnaB is essential to lock formation as strand separation is required before the C6 base can twist out of the helix; this base is located on the displaced strand, which explains why strand displacement by DnaB is halted. This C6 binds near the α4 helix, in or near the DNA-binding channel. Mulcair et al. also showed that while this conformational change occurs at the non-permissive end of the complex when "locked", the interdomain and permissive face remain in a similar conformation to the non-locked complex.
is a particularly important residue as it exists as its conjugate acid in the locked complex, forming hydrogen bonds with C6. Other residues - for example Phe140 and Gly149 - are also strictly conserved amongst different species' Tus proteins, and many of the conserved residues among different Ter sites make base-specific contacts with Tus.
The locked Tus-Ter complex is the most stable known monomeric DNA binding protein with a double-stranded sequence-specific recognition sequence, with a reported half life of 550min (Mulcair et al., 2006). The formation of a large hydrogen-bond network is critical to sequence recognition and the stability of the twisted β-strands lying across the major groove.[11]
Just when we thought we had a nice, elegant theory...
In 2008, Bastia et al. proposed that Tus is actually a polar antitranslocase, and that an AT-GC transversion at position 6 did not affect DnaB-translocation in vitro. They suggested that the base-flipping of C6 functions as a fail-safe mechanism, and that the replication fork is halted primarily by Tus-DnaB and Tus-Ter interactions.[12]
Comparison of RTP and Tus
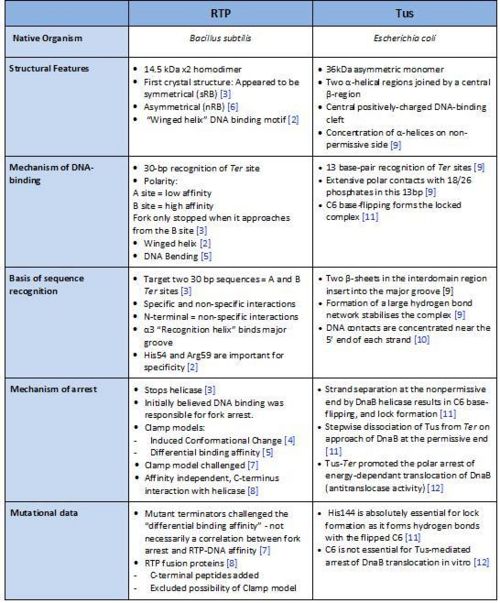
A summary of the different features of fork arrest proteins RTP and Tus
Replication Fork Termination: The Future of Discoveries
Perhaps surprisingly, replication fork termination has also been identified in specific regions of eukaryotes chromosoms. In the yeast Saccharomyces cerevisiae the terminator protein Fob1p has been shown to arrest the replication fork at Ter sites in non-transcribed spacer regions of rDNA; while in Schizosaccharomyes pombe polar replication termination by RTS1 has been shown to control the direction of replication of the mat1 mating locus, enabling the yeast to alternate between mating types. This provides evidence that the replication termination process has been adapted by a wide range of organisms and is able to perform a variety of functions, opening up an exciting new field of research.[13]
3D structures of replication terminator protein
Replication Termination Protein
References
- ↑ Wake, RG and King, GF (1997) A tale of two terminators: crystal structures sharpen the debate on DNA replication fork arrest mechanisms. Structure 5: 1-5.
- ↑ Wilce JA, Vivian JP, Hastings AF, Otting G, Folmer RHA, Duggin IG, Wake RG, Wilce MCJ (2001) Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology 8: 206-210.
- ↑ Bussiere DE, Bastia D, White SW (1995) Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell 80(4): 651-60.
- ↑ Smith MT, de Vries CJ, Langley DB, King GF, Wake RG (1996) The Bacillis subtilis DNA Replication Terminator. Journal of Molecular Biology 260: 54-69.
- ↑ Kralicek, AV, Wilson PK, Ralston GB, Wake RG, King GF (1997) Reorganization of terminator DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex. Nucleic Acids Research 25(3): 590-596.
- ↑ Vivian JP, Porter CJ, Wilce JA, Wilce MCJ (2007) An asymmetric structure of the Bacillus subtilis Replication Terminator Protein in complex with DNA. Journal of Molecular Biology 370: 481-491.
- ↑ Duggin IG, Matthews JM, Dixon, NE, Wake RG, Mackay JP (2004) A Complex Mechanism Determines Polarity of DNA Replication Fork Arrest by the Replication Terminator Complex of Bacillus subtilis. The Journal of Biological Chemistry 280(13): 13105-13113.
- ↑ Duggin, IG (2006) DNA Replication Fork Arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding. Journal of Molecular Biology 361: 1-6.
- ↑ Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K (1996) Structure of a replication-terminator protein complexed with DNA. Nature 383: 598 - 603.
- ↑ Neylon C, Kralicek AV, Hill TM, Dixon NE (2005) Replication Termination in Escherichia coli: Structure and Antihelicase Activity of the Tus-Ter Complex. Microbiology and Molecular Biology Reviews 69: 501 - 526.
- ↑ Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE (2006) A Molecular Mousetrap Determines Polarity of Termination of DNA Replication in E. coli. Cell 125: 1309 - 1319.
- ↑ Bastia D, Zzaman S, Krings G, Saxena M, Peng XH, Greenberg MM (2008) Replication termination mechanism as revealed by Tus-mediated polar arrest of a sliding helicase. Proceedings of the National Academy of Sciences 105: 12831 - 12836.
- ↑ Kaplan DL, Bastia D (2009) Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72(2): 279-285.





