RNaseA Nobel Prizes
From Proteopedia
(Difference between revisions)
| Line 37: | Line 37: | ||
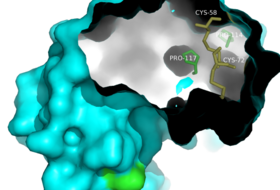
The synthesis of semisynthetic RNasa A clearly exhibits the structure to function relationship that defines proteins. In the RNase A protein, the removal of six C terminal residues, leaving <scene name='Sandbox_Reserved_198/Rnase_1-118/1' target='1'>RNase 1-118</scene>, completely halts enzymatic activity.<ref name="Martin" /> However, a complex of RNase 1-118 with a synthetic polypeptide comprising the C terminal residues <scene name='Sandbox_Reserved_198/Synthetic_component/3' target='1'>111-124</scene> restores enzymatic activity to RNase A. Upon the addition of the synthetic chain, the <scene name='Sandbox_Reserved_198/Interface/7' target='1'>semisynthetic enzyme</scene> <scene name='Sandbox_Reserved_198/Interface/8' target='1'>(Zoom)</scene> adopts a structure that closely resembles that of <scene name='Sandbox_Reserved_198/Wild_type/1' target='1'>Wild Type RNase A</scene><ref name="Martin" />. The restoration of the structure reconstitutes the enzymatic activity of RNase to 98%<ref name="Martin" />. | The synthesis of semisynthetic RNasa A clearly exhibits the structure to function relationship that defines proteins. In the RNase A protein, the removal of six C terminal residues, leaving <scene name='Sandbox_Reserved_198/Rnase_1-118/1' target='1'>RNase 1-118</scene>, completely halts enzymatic activity.<ref name="Martin" /> However, a complex of RNase 1-118 with a synthetic polypeptide comprising the C terminal residues <scene name='Sandbox_Reserved_198/Synthetic_component/3' target='1'>111-124</scene> restores enzymatic activity to RNase A. Upon the addition of the synthetic chain, the <scene name='Sandbox_Reserved_198/Interface/7' target='1'>semisynthetic enzyme</scene> <scene name='Sandbox_Reserved_198/Interface/8' target='1'>(Zoom)</scene> adopts a structure that closely resembles that of <scene name='Sandbox_Reserved_198/Wild_type/1' target='1'>Wild Type RNase A</scene><ref name="Martin" />. The restoration of the structure reconstitutes the enzymatic activity of RNase to 98%<ref name="Martin" />. | ||
| - | <Structure load='1SRN' size='380' frame='true' align='right' caption='Semisynthetic Ribonuclease A: Residues 114-124 are highlighted in the surface representations of the Wild Type, Fully Synthetic, and Semisynthetic enzymes to emphasize similarity in structure. Also, the surface representation of semisynthetic RNase A illustrates the interface between the synthetic analog and the natural enzyme, [[1srn]] ' scene='Sandbox_Reserved_198/Fully_synthetic/4' target='1'/> | ||
| + | <scene name='Sandbox_Reserved_198/Fully_synthetic/4'>Semisynthetic Ribonuclease A: Residues 114-124 are highlighted in the surface representations of the Wild Type, Fully Synthetic, and Semisynthetic enzymes to emphasize similarity in structure.</scene> Also, the surface representation of semisynthetic RNase A illustrates the interface between the synthetic analog and the natural enzyme, [[1srn]]. | ||
The semi-synthetic RNase A comprises of residues 1-118 and the synthetic analog of residues 111-124. The RNase 1-118 was prepared by successive digestion of RNase A pepsin and carboxypeptidase A<ref>PMID: 6615822 </ref>. The synthetic component, RNase 111-124, was prepared by the use of solid-phase peptide synthetic methods, in which the peptide chain was assembled in the stepwise manner while it was attached at one end to a solid support. The peptide chain was extended by repetitive steps of de-protection, neutralization and coupling until the desired sequence was obtained<ref>PMID: 4921569</ref>. It was important that the synthesis proceeds rapidly and in high yields to prevent side reactions or by-products. | The semi-synthetic RNase A comprises of residues 1-118 and the synthetic analog of residues 111-124. The RNase 1-118 was prepared by successive digestion of RNase A pepsin and carboxypeptidase A<ref>PMID: 6615822 </ref>. The synthetic component, RNase 111-124, was prepared by the use of solid-phase peptide synthetic methods, in which the peptide chain was assembled in the stepwise manner while it was attached at one end to a solid support. The peptide chain was extended by repetitive steps of de-protection, neutralization and coupling until the desired sequence was obtained<ref>PMID: 4921569</ref>. It was important that the synthesis proceeds rapidly and in high yields to prevent side reactions or by-products. | ||
| Line 55: | Line 55: | ||
* [[RNase A NMR]] | * [[RNase A NMR]] | ||
* [[RNaseS RNaseB|RNase S and RNase B]] | * [[RNaseS RNaseB|RNase S and RNase B]] | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
==3D structures of ribonuclease== | ==3D structures of ribonuclease== | ||
Revision as of 10:49, 30 July 2013
| |||||||||||
3D structures of ribonuclease
See Also
References
- ↑ 1.0 1.1 Raines RT. Ribonuclease A. Chem Rev. 1998 May 7;98(3):1045-1066. PMID:11848924
- ↑ 'Anfinsen Nobel Lecture' [1]
- ↑ 'Anfinsen Nobel Biography' [2]
- ↑ Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223-30. PMID:4124164
- ↑ 5.0 5.1 'Lehninger A., Nelson D.N, & Cox M.M. (2008) Lehninger Principles of Biochemistry. W. H. Freeman, fifth edition.'
- ↑ 6.0 6.1 6.2 Pearson MA, Karplus PA, Dodge RW, Laity JH, Scheraga HA. Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A. Protein Sci. 1998 May;7(5):1255-8. PMID:9605332 doi:10.1002/pro.5560070522
- ↑ Schultz DA, Friedman AM, White MA, Fox RO. The crystal structure of the cis-proline to glycine variant (P114G) of ribonuclease A. Protein Sci. 2005 Nov;14(11):2862-70. Epub 2005 Sep 30. PMID:16199662 doi:10.1110/ps.051610505
- ↑ Hogan, Dan. ed. Dysfunctional Protein Dynamics Behind Neurological Disease? ScienceDaily.2 Nov. 2009. www.sciencedaily.com [3]
- ↑ 9.0 9.1 9.2 9.3 Merrifield B. "Solid Phase Synthesis", Nobel Lecture, 8 December, 1984.
- ↑ 10.0 10.1 10.2 10.3 Martin PD, Doscher MS, Edwards BF. The refined crystal structure of a fully active semisynthetic ribonuclease at 1.8-A resolution. J Biol Chem. 1987 Nov 25;262(33):15930-8. PMID:3680234
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 Boerema DJ, Tereshko VA, Kent SB. Total synthesis by modern chemical ligation methods and high resolution (1.1 A) X-ray structure of ribonuclease A. Biopolymers. 2008;90(3):278-86. PMID:17610259 doi:10.1002/bip.20800
- ↑ Doscher MS, Martin PD, Edwards BF. Characterization of the histidine proton nuclear magnetic resonances of a semisynthetic ribonuclease. Biochemistry. 1983 Aug 16;22(17):4125-31. PMID:6615822
- ↑ Lin MC. The structural roles of amino acid residues near the carboxyl terminus of bovine pancreatic ribonuclease A. J Biol Chem. 1970 Dec 25;245(24):6726-31. PMID:4921569
External Resources
Student Contributors
Proteopedia Page Contributors and Editors (what is this?)
R. Jeremy Johnson, Wayne Decatur, Angel Herraez, Michal Harel, Eric Martz, Alexander Berchansky, Karsten Theis