We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 126
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='4dki' size=525 side=right scene='4dki'/Com_view/1'> | <StructureSection load='4dki' size=525 side=right scene='4dki'/Com_view/1'> | ||
== Background == | == Background == | ||
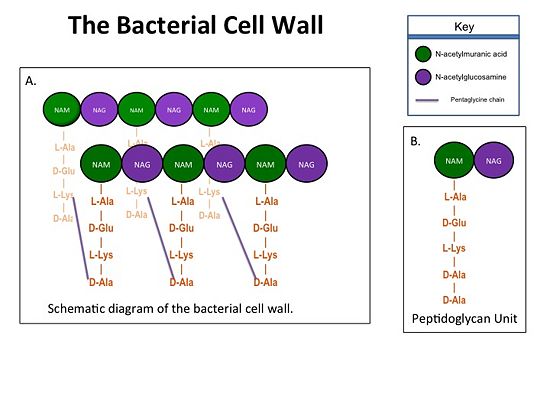
| - | Transpeptidases (TP), also known as penicillin-binding proteins (PBP), catalyze the cross-linking of peptidoglycan polymers during bacterial cell wall synthesis. The natural transpeptidase substrate is the D-Ala-D-Ala peptidoglycan side chain terminus. Beta-lactam (β-lactam) antibiotics, which include penicillins, cephalosporins and carbapenems, bind and irreversibly inhibit transpeptidases by mimicking the D-Ala-D-Ala substrate, resulting in the inhibition of cell wall synthesis and ultimately bacterial cell growth. [[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1. A.Bacterial Cell Wall B.Peptidoglycan with D-Ala-D-Ala substrate |550px]] Overuse and misuse of β-lactams has led to the generation of methicillin- resistant Staphylococcus aureus (MRSA) isolates that have acquired an alternative transpeptidase, PBP2a, which is neither bound nor inhibited by β- lactams. MRSA isolates are resistant to all β-lactams, and are often only susceptible to “last resort antibiotics”, such as vancomycin. Recently, two cephalosporins - <scene name='37/372726/Ceftobiprole/ | + | Transpeptidases (TP), also known as penicillin-binding proteins (PBP), catalyze the cross-linking of peptidoglycan polymers during bacterial cell wall synthesis. The natural transpeptidase substrate is the D-Ala-D-Ala peptidoglycan side chain terminus. Beta-lactam (β-lactam) antibiotics, which include penicillins, cephalosporins and carbapenems, bind and irreversibly inhibit transpeptidases by mimicking the D-Ala-D-Ala substrate, resulting in the inhibition of cell wall synthesis and ultimately bacterial cell growth. [[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1. A.Bacterial Cell Wall B.Peptidoglycan with D-Ala-D-Ala substrate |550px]] Overuse and misuse of β-lactams has led to the generation of methicillin- resistant Staphylococcus aureus (MRSA) isolates that have acquired an alternative transpeptidase, PBP2a, which is neither bound nor inhibited by β- lactams. MRSA isolates are resistant to all β-lactams, and are often only susceptible to “last resort antibiotics”, such as vancomycin. Recently, two cephalosporins - <scene name='37/372726/Ceftobiprole/5'>Ceftobiprole</scene> and ceftaroline - that bind and inhibit PBP2a have been developed. |
<scene name='37/372726/Pbp2a_with_residues/1'>PBP2a and active site residues</scene> | <scene name='37/372726/Pbp2a_with_residues/1'>PBP2a and active site residues</scene> | ||
| Line 13: | Line 13: | ||
== How does Ceftobiprole Work == | == How does Ceftobiprole Work == | ||
| - | <scene name='37/372726/Ceftobiprole/ | + | <scene name='37/372726/Ceftobiprole/5'>Ceftobiprole</scene> is able to inhibit PBP2a because additional chemical groups at <scene name='37/372726/Ceftobiprole/4'>the R2 position of the cephalosporin backbone</scene> are able to interact with additional amino acid residues in PBP2a; specifically <scene name='37/372726/Tyr446_residue/1'>Tyr446</scene> and Met641. As a result of its tighter binding to PBP2a, ceftobiprole is able to more efficiently react with the serine active site residue and therefore inhibit the activity of PBP2a. |
Revision as of 18:27, 31 July 2013
| |||||||||||