We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 123
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
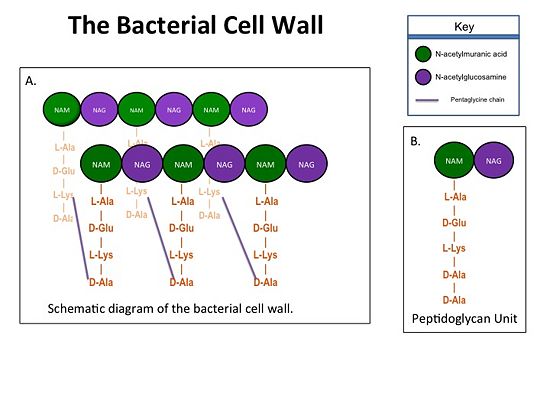
== Catalytic Mechanism of PBP2a == | == Catalytic Mechanism of PBP2a == | ||
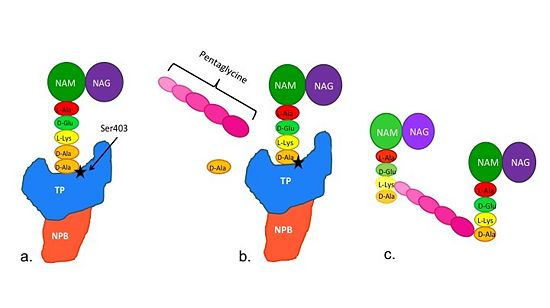
[[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text| |550px]] | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text| |550px]] | ||
| - | (a) The D-Ala-D-Ala side-chain substrate of the peptidoglycan accesses | + | |
| + | (a)The D-Ala-D-Ala side-chain substrate of the peptidoglycan accesses | ||
the active site of the PBP2a. | the active site of the PBP2a. | ||
| - | (b) Ser403 nucleophilically attacks the peptide bond of the terminal | + | (b)Ser403 nucleophilically attacks the peptide bond of the terminal |
D-Ala residues of the substrate. The terminal D-Ala residue then exits | D-Ala residues of the substrate. The terminal D-Ala residue then exits | ||
the active site. The now terminal D-Ala residue forms a covalent bond to Ser403, | the active site. The now terminal D-Ala residue forms a covalent bond to Ser403, | ||
while a crosslinking pentaglycine chain enters the active site. | while a crosslinking pentaglycine chain enters the active site. | ||
| - | (c) A covalent bond forms between the pentaglycine chain and the | + | (c)A covalent bond forms between the pentaglycine chain and the |
terminal D-Ala residue, regenerating the active site serine residue. | terminal D-Ala residue, regenerating the active site serine residue. | ||
Revision as of 17:42, 14 August 2013
| |||||||||||