We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 127
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load= size=475 side='right' scene='36/365380/4dki_cartoon/22'> | <StructureSection load= size=475 side='right' scene='36/365380/4dki_cartoon/22'> | ||
| - | Peptidoglycan | + | Peptidoglycan transpeptidases (TP), also known as penicillin-binding proteins (PBP), are essential for bacterial cell wall synthesis and catalyze the cross-linking of peptidoglycan polymers during bacterial cell wall synthesis. Beta-lactam (β-lactam) antibiotics, which include the penicillins, cephalosporins, carbapenems, and the monobactam aztreonam (Figure 1); bind and irreversibly inhibit the active site of TP. The overuse and misuse of β-lactam antibiotics has led to strains of Staphylococcus aureus (S. aureus) that are resistant to almost all currently available β-lactams and are often only susceptible to so-called “last resort antibiotics”, such as vancomycin. |
| + | |||
| + | |||
== Cell Wall Structure == | == Cell Wall Structure == | ||
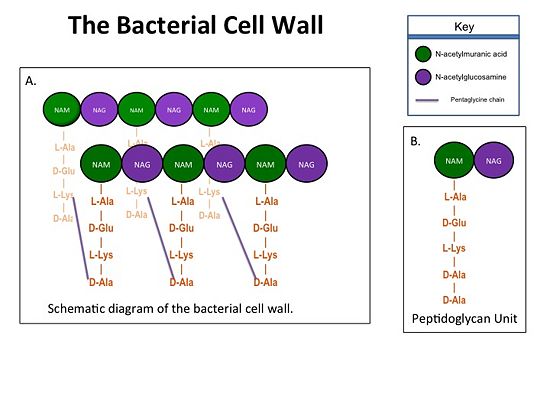
| - | + | The bacterial cell wall is crucial for maintaining the structural integrity of bacteria and protects bacteria from osmotic stress and toxic compounds. The cell wall is composed of peptidoglycan (Figure 2 ) and in Gram positive bacterial species (e.g., S. aureus) is many layers thick, while in Gram negative bacterial species (e.g., Escherichia coli) is only a few layers thick. The difference in the number of peptidoglycan layers accounts for the differential staining of these two groups of organisms. . Peptidoglycan consists of a carbohydrate portion: alternating residues of N-acetylmuranic Acid (NAM) and N-acetylglucosamine (NAG) that polymerize to form long chains, and a protein portion: a pentapeptide chain that terminates with two D-alanine (D-Ala) residues. The pentapeptide chains are covalently bound to each NAM residue Rows of peptidoglycan are cross-linked together with pentaglycine chains to form a "mesh-like" structure. This cross-linking reaction is catalyzed by TPs. | |
| - | The cell wall | + | |
[[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1. A.Bacterial Cell Wall B.Peptidoglycan with D-Ala-D-Ala substrate |550px]] | [[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1. A.Bacterial Cell Wall B.Peptidoglycan with D-Ala-D-Ala substrate |550px]] | ||
| Line 15: | Line 16: | ||
| - | == Catalytic Mechanism of | + | == Catalytic Mechanism of Action of Transpeptidases == |
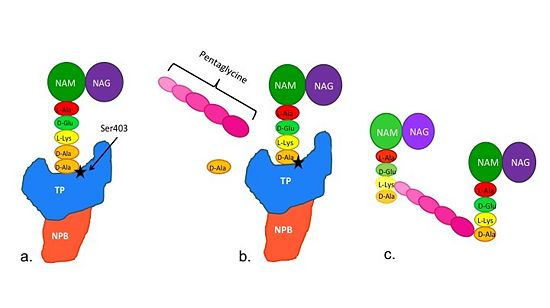
[[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic diagram illustrating the mechanism of action of PBP2a |550px]] | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic diagram illustrating the mechanism of action of PBP2a |550px]] | ||
| + | Note: Schematic is above these statements: | ||
| + | (a)The D-Ala-D-Ala side-chain substrate accesses the TP active site. | ||
| + | (b) The active site serine residue nucleophilically attacks the peptide bond between the terminal D-Ala residues. The terminal D-Ala residue exits the active site, and the remaining D-Ala residue forms a covalent bond with the active site serine residue to form an acyl-TP complex. | ||
| + | (c) Subsequently, a pentaglycine chain enters the TP active site through nucleophillic attack forms a covalent bond with the D-Ala residue formerly bound to the active site serine residue. As a result, the TP active site serine residue is regenerated. The entire process takes approximately 4 milliseconds. | ||
| - | (a)The D-Ala-D-Ala side-chain substrate of the peptidoglycan accesses the active site of the PBP2a. | ||
| - | |||
| - | (b)Ser403 nucleophilically attacks the peptide bond of the terminal D-Ala residues of the substrate. The terminal D-Ala residue then exits the active site. The now terminal D-Ala residue forms a covalent bond to Ser403, while a crosslinking pentaglycine chain enters the active site. | ||
| - | |||
| - | (c)A covalent bond forms between the pentaglycine chain and the terminal D-Ala residue, regenerating the active site serine residue. | ||
| - | + | == Mechanism of Action of β-Lactam Antibiotics == | |
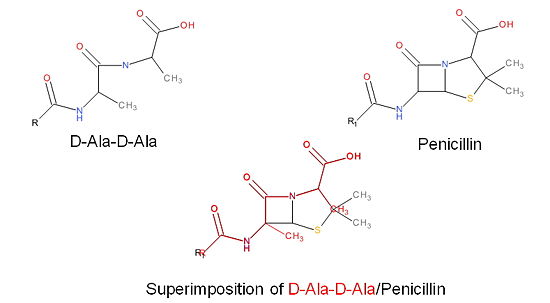
| - | + | The β-lactam antibiotics inhibit bacterial growth by irreversibly inhibiting TPs and, therefore, bacterial cell wall synthesis. Specifically, β-lactams are molecular mimics of a portion of the peptidoglycan polymer, namely the D-Ala-D-Ala moiety, which is the normal TP enzymatic substrate (Figure 4). As a result, bacterial TP enzymes are "tricked" into reacting with β-lactams. Additionally, the β-lactams are very reactive molecules due to their β-lactam ring, and readily react with the TP active site serine residue and sterically block the active site preventing the entry of nucleophiles [http://en.wikipedia.org/wiki/Nucleophile] that regenerate the active site serine residue such as the pentaglycine chain or water. | |
| - | The β-lactam antibiotics inhibit bacterial growth by inhibiting PBPs and ultimately cell wall | ||
| - | synthesis. Specifically, β-lactams are molecular mimics of D-Ala-D-Ala, which is the normal | ||
| - | substrate of PBPs. Nucleophillic attack [http://en.wikipedia.org/wiki/Nucleophile] of the β-lactam results in the PBP being irreversibly inhibited by the β-lactam. As a result, the synthesis of the cell wall is inhibited which leads to cell lysis. | ||
[[Image:Structures on penicillin and b lactam.jpg|thumb|alt= Alt text|Figure 3. Mechanism of action of β-lactams. A. Structure of a β-lactam (penicillin) showing the amide, carboxyl, and β-lactam ring groups β-lactam ring groups. B. Structure of the D-Ala-D-Ala substrate. C. Overlay of the D-Ala-D-Ala substrate in red with penicillin demonstrating molecular mimicry.|550 px]] | [[Image:Structures on penicillin and b lactam.jpg|thumb|alt= Alt text|Figure 3. Mechanism of action of β-lactams. A. Structure of a β-lactam (penicillin) showing the amide, carboxyl, and β-lactam ring groups β-lactam ring groups. B. Structure of the D-Ala-D-Ala substrate. C. Overlay of the D-Ala-D-Ala substrate in red with penicillin demonstrating molecular mimicry.|550 px]] | ||
Revision as of 21:18, 11 September 2013
| |||||||||||