We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 128
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
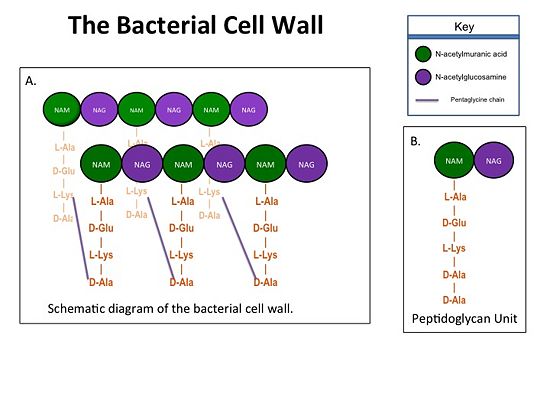
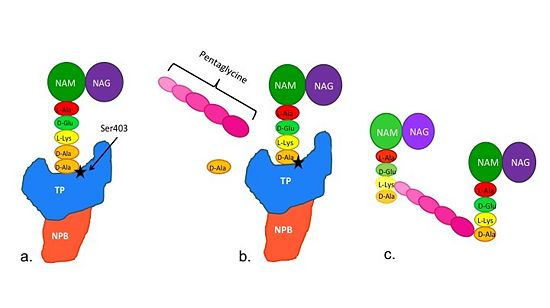
==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ||
(a)The D-Ala side-chain substrate accesses the TP active site. | (a)The D-Ala side-chain substrate accesses the TP active site. | ||
| + | |||
(b)The active site serine residue nucleophilically attacks the peptide bond between the terminal D-Ala residues. Terminal D-Ala residue exits the active site, and the remaining D-Ala residue forms a covalent bond with the active site serine residue to form an acyl-TP complex. | (b)The active site serine residue nucleophilically attacks the peptide bond between the terminal D-Ala residues. Terminal D-Ala residue exits the active site, and the remaining D-Ala residue forms a covalent bond with the active site serine residue to form an acyl-TP complex. | ||
| + | |||
(c)Subsequently, a pentaglycine chain enters the TP active site through nucleophillic attack forms a covalent bond with the D-Ala formerly bound to the active site serine residue. As a result, the TP site serine residue is regenerated. The entire process takes approximately 4 seconds. | (c)Subsequently, a pentaglycine chain enters the TP active site through nucleophillic attack forms a covalent bond with the D-Ala formerly bound to the active site serine residue. As a result, the TP site serine residue is regenerated. The entire process takes approximately 4 seconds. | ||
[[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic showing Catalytic Mechanism of PBP2a |550px]] | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic showing Catalytic Mechanism of PBP2a |550px]] | ||
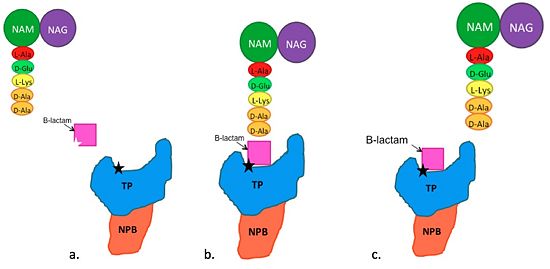
| - | ==='''Mechanism of Action of B-Lactam | + | ==='''Mechanism of Action of B-Lactam Antibiotics'''=== |
The B-Lactam antibiotics inhibit bacterial cell growth by irreversibly inhibiting TP's and, therefore, bacterial cell wall sythesis. Specifically, B-Lactams are molecular mimics of a portion of the peptidoglycan polymer, namely the D-Ala-D-Ala moiety, which is the normal TP enzymatic substrate(Figure 4). As a result, bacterial TP enzymes are "tricked" into reacting with B-Lactams. Additionally, the B-Lactams are very reactive molecules due to their B-lactam ring, and readily react with the TP active site serine residue and sterically block the active site preventing the entry of nucleophiles that regenerate the active site serine residue such as the pentaglycine chain or water. | The B-Lactam antibiotics inhibit bacterial cell growth by irreversibly inhibiting TP's and, therefore, bacterial cell wall sythesis. Specifically, B-Lactams are molecular mimics of a portion of the peptidoglycan polymer, namely the D-Ala-D-Ala moiety, which is the normal TP enzymatic substrate(Figure 4). As a result, bacterial TP enzymes are "tricked" into reacting with B-Lactams. Additionally, the B-Lactams are very reactive molecules due to their B-lactam ring, and readily react with the TP active site serine residue and sterically block the active site preventing the entry of nucleophiles that regenerate the active site serine residue such as the pentaglycine chain or water. | ||
[[Image:BLactamSchematic2.jpg|thumb|alt= Alt text|550px]] | [[Image:BLactamSchematic2.jpg|thumb|alt= Alt text|550px]] | ||
Revision as of 21:10, 16 October 2013
| |||||||||||