Superoxide Dismutase
From Proteopedia
| Line 1: | Line 1: | ||
<StructureSection load='Combo.pdb' size='450' side='right' scene='Superoxide_Dismutase/Opening_sod/2' caption=''> | <StructureSection load='Combo.pdb' size='450' side='right' scene='Superoxide_Dismutase/Opening_sod/2' caption=''> | ||
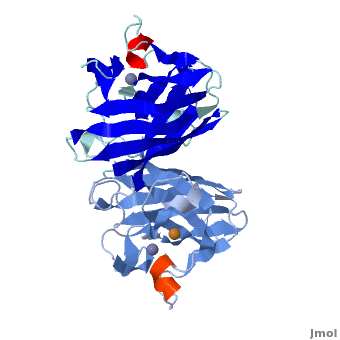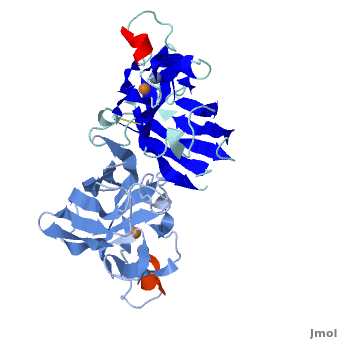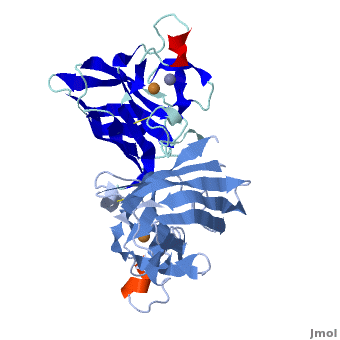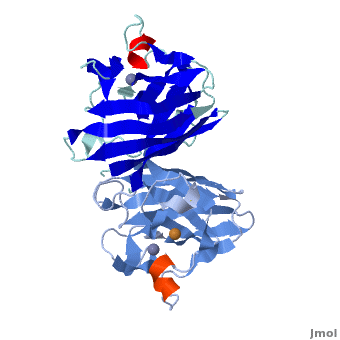
[[Image:1spd.png|200px|left|thumb|Crystal Structure of Human Superoxide Dismutase, [[1spd]]]] | [[Image:1spd.png|200px|left|thumb|Crystal Structure of Human Superoxide Dismutase, [[1spd]]]] | ||
| - | + | {{clear}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
'''Superoxide dismutase (SOD)''' are a group of antioxidant enzymes which catalyze the dismutation of superoxide to oxygen and hydrogen peroxide. SODs are critical antioxidative proteins, protecting the host from oxidative damage. See also<br /> | '''Superoxide dismutase (SOD)''' are a group of antioxidant enzymes which catalyze the dismutation of superoxide to oxygen and hydrogen peroxide. SODs are critical antioxidative proteins, protecting the host from oxidative damage. See also<br /> | ||
* [[Molecular Playground/ Copper-Zinc Superoxide Dismutase]]<br /> | * [[Molecular Playground/ Copper-Zinc Superoxide Dismutase]]<br /> | ||
| Line 38: | Line 8: | ||
<br /> | <br /> | ||
| - | + | __NOTOC__ | |
==Types of SOD:== | ==Types of SOD:== | ||
| Line 50: | Line 20: | ||
==Biochemistry== | ==Biochemistry== | ||
| - | Superoxide is a highly reactive oxygen species and is a major source of oxidative stress in the body, reacting with cellular targets, often causing oxidative damage. <ref>PMID:7493016</ref> SOD protects the body by safely metabolizing the superoxide into unreactive oxygen and hydrogen peroxide. Experiments conducted with knockout mice unable to produce SOD develop widespread oxidative damage and hepatocarcinogenesis and exhibit a greatly reduced lifespan. <ref> PMID:15531919</ref> In humans, mutations to SOD1, one of 3 types found in the human body, can cause familial ALS, a motor neuron disease better known as Lou Gherig’s Disease <ref> PMID:10970056</ref>, and has also been linked to Down’s Syndrome<ref>PMID:7999984</ref>. | + | Superoxide is a highly reactive oxygen species and is a major source of oxidative stress in the body, reacting with cellular targets, often causing oxidative damage. <ref>PMID:7493016</ref> SOD protects the body by safely metabolizing the superoxide into unreactive oxygen and hydrogen peroxide. Experiments conducted with knockout mice unable to produce SOD develop widespread oxidative damage and hepatocarcinogenesis and exhibit a greatly reduced lifespan. <ref> PMID:15531919</ref> In humans, mutations to SOD1, one of 3 types found in the human body, can cause familial ALS, a motor neuron disease better known as Lou Gherig’s Disease <ref> PMID:10970056</ref>, and has also been linked to Down’s Syndrome<ref>PMID:7999984</ref>. |
| + | |||
| + | === Carbon monoxide binding to the heme group at the dimeric interface modulates structure and copper accessibility in the Cu,Zn superoxide dismutase from ''Haemophilus ducreyi'': in silico and in vitro evidences <ref>DOI 10.1080/07391102.2012.680028</ref>=== | ||
| + | <scene name='Journal:JBSD:9/Cv/5'>Superoxide dismutases</scene> (SODs) are metalloenzymes playing a vital role in the defense mechanism against the oxidative stress; they catalyze the dismutation of superoxide, the one-electron reduction product of oxygen, to hydrogen peroxide and molecular oxygen, thus protecting living organism from oxidative lethality (<scene name='Journal:JBSD:9/Cv/7'>conserved SODs regions</scene> among prokaryotes and eukaryotic organisms are highlighted in different colors: <font color='crimson'><b>SS subloop (residues Glu73-Gly93) in crimson</b></font>; (<span style="color:salmon;background-color:black;font-weight:bold;">Zn subloop (residues Gly94-Ala119) in salmon</span>; (<span style="color:violet;background-color:black;font-weight:bold;">Greek key loop (residues Pro135-Gly145) in violet color</span>; (<span style="color:pink;background-color:black;font-weight:bold;">7,8 loop (residues Ala152-Pro169) in pink color)</span>. ''Haemophilus ducreyi'', the causative agent of the sexually transmitted human genital ulcerative disease known as chancroid, expresses one of the most interesting examples of bacterial Cu,Zn SOD (#HdSOD) with the unique feature of binding a heme molecule at the interface between the two subunits asymmetrically bound by residues <scene name='Journal:JBSD:9/Cv/11'>His64 and His124 of subunits A and B</scene>, respectively (<span style="color:lime;background-color:black;font-weight:bold;">His64 and His124 are colored green</span> and <span style="color:yellow;background-color:black;font-weight:bold;">heme molecule is in yellow)</span>. The heme molecule proved to be able to bind small gaseous ligands, such as nitric oxide or carbon monoxide, as a sixth ligand thus displacing the distal histidine. In this study the structural and dynamic response of HdSOD to the <scene name='Journal:JBSD:9/Cv/12'>binding of CO to heme</scene> (<font color='magenta'><b>the carbon atom of CO colored in magenta</b></font> and <font color='red'><b>the oxigen one is in red</b></font>) was studied by means of a combinations of Molecular Dynamics Simulation and X-Ray absorbtion spectroscopy and hypothesis formulated were further confirmed by in vitro experiments. All together the collected results evidenced that binding of the CO molecule produces a strong reduction in the asymmetric fluctuations of the two subunits and long range effects of the heme group on the <scene name='Journal:JBSD:9/Cv/13'>Cu active site structure</scene> (<span style="color:cyan;background-color:black;font-weight:bold;">active site residues: His70, His72, His95, His104, His113, Asp 116 and His 151 are in cyan</span>, <font color='gray'><b>Zn is in gray</b></font> and <span style="color:darkgoldenrod;background-color:black;font-weight:bold;">Cu is in darkgoldenrod</span>) which becomes more easily accessible to the solvent thus causing an increase in copper dismutasic activity. Based on this picture, we suggest a role of HdSOD as a heme-based-sensor protein in which conformational changes, triggered by the heme group, could help ''Haemophilus ducreyi'' to adapt at fluctuating levels of gaseous molecules, such as carbon monoxide or nitric oxide: in the presence of these gasses, the HdSOD would be able to increase its superoxide dismutation activity to subtract superoxide substrate and to prevent the formation of the dangerous peroxynitrite, the molecule generated by the reaction of nitric oxide with superoxide, which is even more harmful to the cell than superoxide. Further investigation to validate this attractive hypothesis is thus desirable. | ||
==Additional Resources== | ==Additional Resources== | ||
See: [[Copper, Zinc Superoxide Dismutase]] <br/> | See: [[Copper, Zinc Superoxide Dismutase]] <br/> | ||
Revision as of 09:16, 13 November 2013
| |||||||||||
3D Structures of SOD
Updated on 13-November-2013
Metal-free apo SOD
3hog – SOD – tomato
1p7g – SOD – Pyrobaculum aerophilum
1ozt, 3gzp, 3gzq, 3ecv, 3ecw, 2gbu - hSOD (mutant) - human
3ecu, 3k91 – hSOD
1rk7 - hSOD (mutant) – NMR
3kbe – CeSOD – Caenorhabditis elegans
1t6i, 1t6q – ScSOD - Streptomyces coelicolor
3ak1 - ApSOD - Aeropyrum pernix
Cu-Zn-SOD
2jlp, 2v0a, 2c9v, 2c9u, 2c9s, 1pu0, 1hl5, 1sos, 1spd, 3kh3, 3kh4, 3re0, 3t5w, 4b3e- hSOD +Zn+Cu - human
3h2p, 3h2q, 3h2r, 3hff, 3gqf, 2nnx, 1p1v, 1oez, 2xjl, 3qqd, 4bd4, 4bcz, 4bcy – hSOD (mutant) +Zn
2af2, 1kmg - hSOD (mutant) +Zn – NMR
1l3n, 1dsw, 1ba9, 2lu5 – hSOD (mutant) +Zn+Cu - NMR
2wyz - hSOD (mutant) +Zn+Cu+UMP
2wz0 - hSOD (mutant) +Zn+Cu+aniline
2wz5 - hSOD (mutant) +Zn+Cu+methionine
2wz6 - hSOD (mutant) +Zn+Cu+quinazoline
2r27 - hSOD +Cu
1hl4 - hSOD +Zn
2wko, 3gzo, 2zkw, 2zkx, 2zky, 3cqp, 3cqq, 2vr6, 2vr7, 2vr8, 2gbt, 2gbv, 1uxl, 1uxm, 1ptz, 1ozu, 1n18, 1n19, 1fun, 1mfm, 1azv, 2wyt, 2xjk – hSOD (mutant) +Zn+Cu
4a7g, 4a7q - hSOD (mutant) +Zn+Cu + quinazoline derivative
4a7s - hSOD (mutant) +Zn+Cu + uridine derivative
4a7t - hSOD (mutant) +Zn+Cu + isoproterenol
4a7u - hSOD (mutant) +Zn+Cu + adrenaline
4a7v - hSOD (mutant) +Zn+Cu + dopamine
3l9e - smSOD +Zn – silk moth
3l9y - smSOD +Zn+Cu
2wn0, 2wwn, 2wwo – YpSOD+Zn+Cu – Yersinia pseudotuberculosis
2wn1 - YpSOD+Zn+Cu+N3
2z7u, 2z7w, 2z7y, 2z7z, 2aeo, 1q0e, 1e9p, 1e9q, 1cb4, 1cbj, 1sxn, 1sxa, 1sxb, 1sxc, 1spd, 1sda, 3sod, 2sod, 2zow, 3hw7 – cSOD+Zn+Cu - cow
1cob – cSOD+Cu+Co
1sxs - cSOD +Zn+Cu+SCN
1sxz - cSOD +Zn+Cu+N3
1e9o – cSOD+Cu
3f7k - ApSOD +Zn+Cu+H2O2 – Alvinella pompejana
3f7l - ApSOD +Zn+Cu
2k4w – SeSOD (mutant) +Zn+Cu – Salmonella enterica – NMR
1eqw - SOD +Zn+Cu – Salmonella typhimurium
3ce1 - SOD +Zn+Cu – Cryptococcus liquefaciens
2q2l - SOD +Zn – Potentilla atrosanguinea
2aqm - SOD +Zn+Cu – Brucella abortus
2aps - SOD +Zn+Cu – Actinobacillus pleuropneumoniae
2aqn - NmSOD +Zn+Cu – Neisseria meningitides
2aqp, 2aqq, 2aqr, 2aqs, 2aqt – NmSOD (mutant) +Zn+Cu
1z9n - HdSOD +Zn+Cu+haem – Haemophilus ducreyi
1z9p - HdSOD +Zn+Cu
1to4, 1to5 - SOD +Zn+Cu – Schistosoma mansoni
1pzs - MtSOD +Cu – Mycobacterium tuberculosis
1oaj, 1bzo, 1yai - PlSOD +Zn+Cu – Photobacterium leiognathi
1oal, 1ib5, 1ibb, 1ibd, 1ibf, 1ibh – PlSOD (mutant) +Zn+Cu
1f18, 1f1a, 1f1d, 1b4t – ySOD (mutant) +Zn+Cu – yeast
1f1g, 1b4l, 2jcw, 1sdy - ySOD +Zn+Cu
1yaz - ySOD +Zn+Cu+N3
1jk9 – ySOD+Zn+Cu chaperone for SOD
1eso - EcSOD +Zn+Cu - Escherichia coli
1yso - EcSOD +Zn+Cu
1srd – SOD+MZn+Cu –Spinach
3gtt – mSOD+Zn – mouse
3gtv, 3ltv – mSOD/hSOD+Zn chimera
3kbf – CeSOD+Zn+Cu
3mkg, 3km1, 3km2 – tSOD + Zn - tomato
3mnd – SOD + Zn + Cu – Pig tapeworm
3pu7, 3s0p - tSOD + Zn + Cu
Mn-SOD
Human Mn-SOD is discussed in the article on Nitrotyrosine.
3k9s – EcSOD+H2O2
1d5n, 1vew, 3ot7 - EcSOD+Mn
1zlz, 1ixb, 1ix9, 1en4, 1en5, 1en6, 1i08, 1i0h – EcSOD (mutant)+Mn
3dc5, 3dc6 – SOD+Mn – Caenorhabditis elegans
3bfr, 1jcv, 3lsu, 3rn4 – ySOD+Mn
4e4e, 4f6e - ySOD (mutant)+Mn
2qka, 2adp, 2adq, 1xdc, 1xil, 1n0j, 1luv, 1msd – hSOD+Mn
2qkc, 3c3s, 3c3t, 2p4k, 1zsp, 1zte, 1zuq, 2gds, 1szx, 1pl4, 1pm9, 1n0n, 1luw, 1ja8, 1em1, 1ap5, 1ap6, 1qnm, 1var – hSOD (mutant)+Mn
2rcv – SOD+Mn – Bacillus subtilis
1xre, 1xuq – SOD+Mn – Bacillus anthracis
2aw9, 2cdy, 2ce4, 3kky – DrSOD+Mn – Deinococcus radiodurans
2a03 – SOD+Mn+Zn – Plasmodium berghei
1jr9 - SOD +Zn+Mn – Virgibacillus halodenitrificans
1gv3 – SOD+Mn – Anabaena
1kkc – SOD+Mn – Aspergillus fumigatus
1xso – SOD+Mn – Xenopus laevis
1mng, 3mds – TtSOD+Mn – Thermus thermophilus
3evk – SOD+Mn – Pyrobaculum aerophilum
1ar4, 1ar5 – PfSOD+Mn - Propionibacterium freudenreichii
3ak2 - ApSOD+Mn
Fe-SOD
3js4 – SOD+Fe – Anaplasma phagocytuphilum
1mmm – EcSOD+Fe
1y67 – DrSOD+Fe
3esf – SOD (mutant)+Fe – Trypanosoma brucei
2gpc, 4dvh – SOD+Fe – Trypanosoma cruzi
3h1s – SOD+Fe – Francisella tularensis
2w7w – SOD+Fe – Aliivibrio salmonicida
3cei – SOD+Fe – Helicobacter pylori
2goj, 2bpi – PfSOD+Fe – Plasmodium falciparum
2awp – SOD+ion – Plasmodium knowlesi
2bkb, 2nyb, 1za5 – EcSOD (mutant)+Fe
2cw2, 2cw3 – SOD+Fe – Perkinsus marinus
1wb7, 1wb8 – SOD (mutant)+Fe – Sulfolobus solfataricus
1unf – SOD+Fe – Vigna unguiculata
1uer, 1ues, 1qnn – SOD+Fe – Porphyromonas gingivalis
1my6 – SOD+Fe – Thermosynechococcus elongates
1ma1 – SOD+Fe – Methanothermobacter thermautotrophicus
1gn2, 1gn3, 1gn4, 1gn6 – MtSOD (mutant)+Fe
1ids – MtSOD+Fe
1dt0, 3sdp – SOD+Fe – Pseudomonas putida
1b06 – SOD+Fe – Sulfolobus acidocaldarius
1bs3 – PfSOD+Fe+F
1bsm, 1bt8 – PfSOD+Fe
1avm – PfSOD+Fe+N3
1coj – SOD+Fe – Aquifex pyrophilus
1isa, 1isb, 1isc – EcSOD+Fe
3lio, 3ljt, 3lj9, 3ljf – SOD+Fe
3ak3 - ApSOD+Fe
3tqj – SOD + Fe – Coxiella burneti
4f2n - SOD + Fe – Leishmania major
3tjt - SOD + Fe – Clostridium difficile
4ffk – SOD + Fe – Acidilobus saccharovorans
Ni-SOD
3g4x, 3g4z, 3g50, 1t6u – ScSOD (mutant)+Ni
1q0d, 1q0f, 1q0g, 1q0k, 1q0m – SOD+Ni – Streptomyces seoulensis
Zn-SOD
4k2w – SOD+ Zn – Babesia bovis
References
- ↑ McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969 Nov 25;244(22):6049-55. PMID:5389100
- ↑ McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968-1988). Free Radic Biol Med. 1988;5(5-6):363-9. PMID:2855736
- ↑ Richardson J, Thomas KA, Rubin BH, Richardson DC. Crystal structure of bovine Cu,Zn superoxide dismutase at 3 A resolution: chain tracing and metal ligands. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1349-53. PMID:1055410
- ↑ Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982 Sep 15;160(2):181-217. PMID:7175933
- ↑ Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995 Dec;11(4):376-81. PMID:7493016 doi:http://dx.doi.org/10.1038/ng1295-376
- ↑ Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005 Jan 13;24(3):367-80. PMID:15531919
- ↑ Al-Chalabi A, Leigh PN. Recent advances in amyotrophic lateral sclerosis. Curr Opin Neurol. 2000 Aug;13(4):397-405. PMID:10970056
- ↑ Groner Y, Elroy-Stein O, Avraham KB, Schickler M, Knobler H, Minc-Golomb D, Bar-Peled O, Yarom R, Rotshenker S. Cell damage by excess CuZnSOD and Down's syndrome. Biomed Pharmacother. 1994;48(5-6):231-40. PMID:7999984
- ↑ Chillemi G, De Santis S, Falconi M, Mancini G, Migliorati V, Battistoni A, Pacello F, Desideri A, D'Angelo P. Carbon monoxide binding to the heme group at the dimeric interface modulates structure and copper accessibility in the Cu,Zn superoxide dismutase from Haemophilus ducreyi: in silico and in vitro evidences. J Biomol Struct Dyn. 2012 Jul;30(3):269-79. Epub 2012 Jun 11. PMID:22686457 doi:10.1080/07391102.2012.680028
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Alexander Berchansky, Eric Martz, Joel L. Sussman, Jane S. Richardson, Jaime Prilusky