User:Andrew Wills/Sandbox 1
From Proteopedia
| Line 13: | Line 13: | ||
==Key Structures== | ==Key Structures== | ||
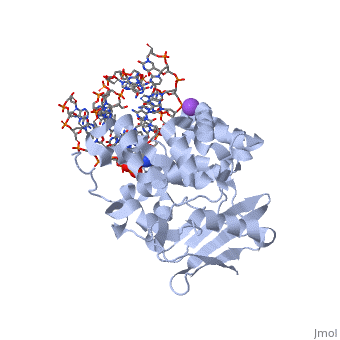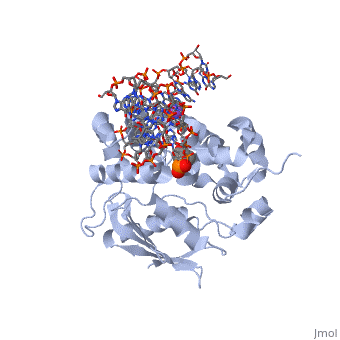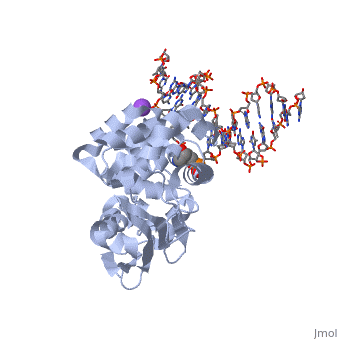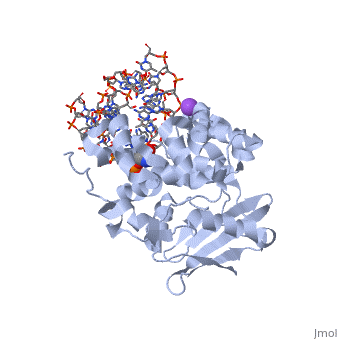
AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms. The first domain (residues 1-112) is the N-terminal domain that is composed of a five stranded antiparallel beta sheet and two alpha helices. The second domain (residues 113-230) contains seven alpha helices that create a hydrophobic core. The third domain (resudues 231-282) is the C-terminal domain that contains a bundle of three alpha helices (Moe). (Labahn) | AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms. The first domain (residues 1-112) is the N-terminal domain that is composed of a five stranded antiparallel beta sheet and two alpha helices. The second domain (residues 113-230) contains seven alpha helices that create a hydrophobic core. The third domain (resudues 231-282) is the C-terminal domain that contains a bundle of three alpha helices (Moe). (Labahn) | ||
| - | AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop (Moe). In AlkA, the <scene name='56/566536/Hhh_domain/2'> | + | AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop (Moe). In AlkA, the <scene name='56/566536/Hhh_domain/2'>HhH</scene> domain is composed of residues 202-227 and is responsible for binding the damaged DNA by van der Waals interactions, a few hydrogen bonds, and metal ion interactions. |
==Active Sites and Mechanism== | ==Active Sites and Mechanism== | ||
Revision as of 02:40, 4 November 2013
| |||||||||
| 1diz, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | DNA-3-methyladenine glycosylase II, with EC number 3.2.2.21 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
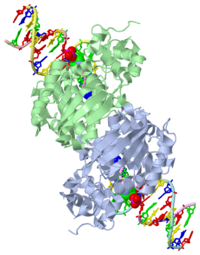
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
General Function
1diz Escherichia coli 3 methyladenine DNA glycosylase II (AlkA) is a DNA repair enzyme that initiates base excision repair for the removal of alkylated bases. Aflatoxin B1 is an example of a toxin that attacks guanine and adenine at their N-7 atom to form alkylated bases (stryker), which prevent regulatory proteins from binding to DNA and blocks replicative polymerases (Hollis). AlkA initiates base excision repair by first locating and binding to the alkylated DNA. It then flips the affected base out of the DNA double helix and into the active site of the enzyme. Once in the active site, AlkA hydrolyzes the glycosidic bond to release the damaged base and leave the sugar phosphate backbone intact. This creates the AP site that is either devoid of a purine or pyridine. The AP site signals to other base excision repair enzymes to insert an undamaged nucleotide based on the undamaged complementary strand and seal the DNA.
| |||||||||||
See Also
Reference
- Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000 Feb 15;19(4):758-66. PMID:10675345 doi:http://dx.doi.org/10.1093/emboj/19.4.758