We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ku protein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
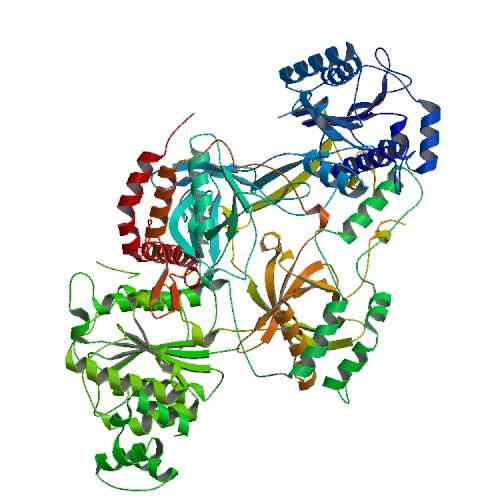
<StructureSection load='1JEY' size='475' side='right' caption='Structure of the Ku heterodimer bound to DNA (PDB entry [[1JEY]])' scene=''> | <StructureSection load='1JEY' size='475' side='right' caption='Structure of the Ku heterodimer bound to DNA (PDB entry [[1JEY]])' scene=''> | ||
| - | |||
== Overview == | == Overview == | ||
The '''Ku protein''' binds to the ends of double-strand breaks and it is required in DNA-repair for non-homologous end joining. The eukaryotic Ku protein is a | The '''Ku protein''' binds to the ends of double-strand breaks and it is required in DNA-repair for non-homologous end joining. The eukaryotic Ku protein is a | ||
| Line 15: | Line 14: | ||
'''Crystal structure of Ku Heterodimer unbound to DNA''' <ref> PMID: 11493912</ref> | '''Crystal structure of Ku Heterodimer unbound to DNA''' <ref> PMID: 11493912</ref> | ||
| - | [[Image:1jey.jpg]] | ||
| - | |||
| - | </StructureSection> | ||
== Structure == | == Structure == | ||
| Line 27: | Line 23: | ||
<scene name='56/567269/Ku70_subunit/3'>Ku70/80 subunits</scene> | <scene name='56/567269/Ku70_subunit/3'>Ku70/80 subunits</scene> | ||
Consisting of three domains (<scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene>, <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene>, <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene>), the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> dimerizes with the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> to form the protein <ref> PMID: 11493912</ref>. Unlike other DNA binding proteins, the Ku protein is asymmetrical from the differences between the Ku70 and Ku80 subunits. This asymmetry leads to different favorable locations for DNA based on major and minor grooves <ref> PMID: 11493912</ref>. The <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> is angled closer to <scene name='56/567269/Bound_dna/3'>DNA</scene> at the double strand break, providing protectiion and interaction with its domains (SOURCE #2). In contrast, the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> associates with <scene name='56/567269/Bound_dna/3'>DNA</scene> away from the free end <ref> PMID: 11493912</ref>. Once a homodimer, the protein has diverged into two domains that are now 15% similar in residues (SOURCE #3). | Consisting of three domains (<scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene>, <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene>, <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene>), the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> dimerizes with the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> to form the protein <ref> PMID: 11493912</ref>. Unlike other DNA binding proteins, the Ku protein is asymmetrical from the differences between the Ku70 and Ku80 subunits. This asymmetry leads to different favorable locations for DNA based on major and minor grooves <ref> PMID: 11493912</ref>. The <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> is angled closer to <scene name='56/567269/Bound_dna/3'>DNA</scene> at the double strand break, providing protectiion and interaction with its domains (SOURCE #2). In contrast, the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> associates with <scene name='56/567269/Bound_dna/3'>DNA</scene> away from the free end <ref> PMID: 11493912</ref>. Once a homodimer, the protein has diverged into two domains that are now 15% similar in residues (SOURCE #3). | ||
| - | |||
=== α/β-Domain === | === α/β-Domain === | ||
| - | + | ||
Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in <scene name='56/567269/Bound_dna/3'>DNA</scene> <ref> PMID: 11493912</ref>. In terms of protein structure, the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> contributes little to the dimer interface between the subunits. The C terminus of the domain can be bound to other repair molecules, using <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> as a scaffold <ref> PMID: 11493912</ref>. | Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in <scene name='56/567269/Bound_dna/3'>DNA</scene> <ref> PMID: 11493912</ref>. In terms of protein structure, the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> contributes little to the dimer interface between the subunits. The C terminus of the domain can be bound to other repair molecules, using <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> as a scaffold <ref> PMID: 11493912</ref>. | ||
| + | |||
=== β-barrel === | === β-barrel === | ||
| - | <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> | ||
The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> itself and <scene name='56/567269/Bound_dna/3'>DNA helix</scene>, with each <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> being composed of seven β strands with the majority in antiparallel arrangement <ref> PMID: 11493912</ref>. The quantity of the strands lends the structures to be symmetrical. Both <scene name='56/567269/Ku70_dimer/4'>β-barrels</scene> in the dimer form the base of the cradle by fitting in the grooves of <scene name='56/567269/Bound_dna/3'>DNA</scene>. | The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> itself and <scene name='56/567269/Bound_dna/3'>DNA helix</scene>, with each <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> being composed of seven β strands with the majority in antiparallel arrangement <ref> PMID: 11493912</ref>. The quantity of the strands lends the structures to be symmetrical. Both <scene name='56/567269/Ku70_dimer/4'>β-barrels</scene> in the dimer form the base of the cradle by fitting in the grooves of <scene name='56/567269/Bound_dna/3'>DNA</scene>. | ||
=== C-terminal arm === | === C-terminal arm === | ||
| - | <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> | ||
The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the <scene name='56/567269/Bound_dna/3'>DNA helix</scene> <ref> PMID: 11493912</ref>. As a result, the <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> strengthens the cradle composed of the two β-barrels. | The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the <scene name='56/567269/Bound_dna/3'>DNA helix</scene> <ref> PMID: 11493912</ref>. As a result, the <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> strengthens the cradle composed of the two β-barrels. | ||
=== DNA binding ring === | === DNA binding ring === | ||
| - | <scene name='56/567269/Ku70_dimer/6'>DNA binding ring</scene> | ||
| - | The DNA binding ring on the open end of DNA is associated with the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene>. By binding <scene name='56/567269/Bound_dna/3'>DNA</scene>, Ku realigns the the strands and protects the molecule from degradation and unwanted bonds while NHEJ occurs <ref> PMID: 11493912</ref>. The regulation of the DNA binding ring of Ku is still under research, with data supporting oxidative stress and redox reactions decreasing the association of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> with <scene name='56/567269/Bound_dna/3'>DNA</scene> through alterations in cysteine residues on the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> (NEED SOURCE 3 and 4, NEED SCENE OF CYSTEINES). | + | The <scene name='56/567269/Ku70_dimer/6'>DNA binding ring</scene> on the open end of DNA is associated with the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene>. By binding <scene name='56/567269/Bound_dna/3'>DNA</scene>, Ku realigns the the strands and protects the molecule from degradation and unwanted bonds while NHEJ occurs <ref> PMID: 11493912</ref>. The regulation of the DNA binding ring of Ku is still under research, with data supporting oxidative stress and redox reactions decreasing the association of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> with <scene name='56/567269/Bound_dna/3'>DNA</scene> through alterations in cysteine residues on the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> (NEED SOURCE 3 and 4, NEED SCENE OF CYSTEINES). |
| + | |||
| + | |||
== Function == | == Function == | ||
| Line 54: | Line 49: | ||
| - | + | </StructureSection> | |
== References== | == References== | ||
<references /> | <references /> | ||
Revision as of 20:07, 4 November 2013
| |||||||||||
References
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000