This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Trypsin
From Proteopedia
| Line 4: | Line 4: | ||
{{TOC limit|limit=2}} | {{TOC limit|limit=2}} | ||
[[Image:Tryogen.gif |thumb|left|Trypsinogen]] | [[Image:Tryogen.gif |thumb|left|Trypsinogen]] | ||
| + | {{Clear}} | ||
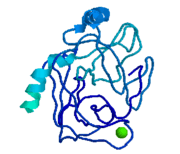
Trypsinogen is the precursor form or zymogen of Trypsin. Zymogens are enzyme precursors that are made and secreted in the lysosome of the cell. Zymogens are not active until they go through a chemical process such as hydrolysis, cleavage, or other cleavages that reveal the active site. The zymogen precursor is necessary in order to prevent the destruction of cellular proteins and to allow the enzyme to be in it's active state only when in appropriate conditions. Trypsinogen is specifically produced in the exocrine cells of the pancreas. There are three isoforms of Trypsinogen that are secreted by the pancreas. The precursor is only activated when it reaches the lumen of the small intestine. This activation occurs through the help of an enteropeptidase and once activated trypsin stimulates the formation of more trypsinogen<ref>Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.</ref>. The structure of Bovine Trypsinogen is shown in the figure to the right <ref> Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html</ref>. | Trypsinogen is the precursor form or zymogen of Trypsin. Zymogens are enzyme precursors that are made and secreted in the lysosome of the cell. Zymogens are not active until they go through a chemical process such as hydrolysis, cleavage, or other cleavages that reveal the active site. The zymogen precursor is necessary in order to prevent the destruction of cellular proteins and to allow the enzyme to be in it's active state only when in appropriate conditions. Trypsinogen is specifically produced in the exocrine cells of the pancreas. There are three isoforms of Trypsinogen that are secreted by the pancreas. The precursor is only activated when it reaches the lumen of the small intestine. This activation occurs through the help of an enteropeptidase and once activated trypsin stimulates the formation of more trypsinogen<ref>Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.</ref>. The structure of Bovine Trypsinogen is shown in the figure to the right <ref> Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html</ref>. | ||
| Line 14: | Line 15: | ||
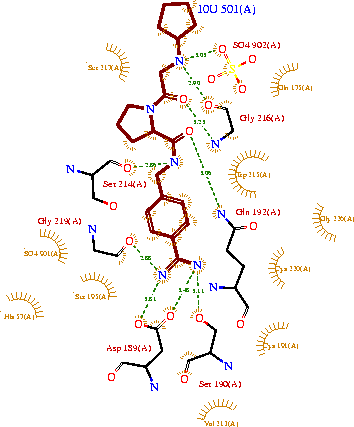
The binding of trypsin to UB-THR 10 somewhat emulates the binding to its specific peptide substrates. The preference for lysine or arginine in trypsin catalysis is due to the composition of the trypsin <scene name='Sandbox_45/Specificitypocketasp189gly216/2'>specificity pocket</scene>. Here (green), Asp 189 and one of two significant glycine backbones, Gly 216, interact with the ligand as they would with Arg or Lys. | The binding of trypsin to UB-THR 10 somewhat emulates the binding to its specific peptide substrates. The preference for lysine or arginine in trypsin catalysis is due to the composition of the trypsin <scene name='Sandbox_45/Specificitypocketasp189gly216/2'>specificity pocket</scene>. Here (green), Asp 189 and one of two significant glycine backbones, Gly 216, interact with the ligand as they would with Arg or Lys. | ||
[[Image:Ligandinteractionstrypsin.gif|thumb|left|upright=2.5|A two-dimensional representation of trypsin binding Ligand UB-THR 10]] | [[Image:Ligandinteractionstrypsin.gif|thumb|left|upright=2.5|A two-dimensional representation of trypsin binding Ligand UB-THR 10]] | ||
| - | + | {{Clear}} | |
The <scene name='Sandbox_45/Ctriadd102h57s195/4'>catalytic triad</scene>; Asp 102, His 57, and Ser 195, shown here in yellow, is positioned near the substrate. The catalytically active histidine and serine side chains are even near an amide bond in UB-THR 10, just like the amide bond broken in peptide hydrolysis. According to FirstGlance in Jmol, there is no bonding of these groups with the ligand, apart from minor van der Waal's interactions with Hist 57. If Ligand UB-Thr 10 were a transition state analog, some covalent connection would exist in addition to hydrogen bonds. UB-THR 10 simulates the substrate, but does not hydrolyze at either of its two amide bonds, likely due to the local cyclic groups atypical of peptide backbones. | The <scene name='Sandbox_45/Ctriadd102h57s195/4'>catalytic triad</scene>; Asp 102, His 57, and Ser 195, shown here in yellow, is positioned near the substrate. The catalytically active histidine and serine side chains are even near an amide bond in UB-THR 10, just like the amide bond broken in peptide hydrolysis. According to FirstGlance in Jmol, there is no bonding of these groups with the ligand, apart from minor van der Waal's interactions with Hist 57. If Ligand UB-Thr 10 were a transition state analog, some covalent connection would exist in addition to hydrogen bonds. UB-THR 10 simulates the substrate, but does not hydrolyze at either of its two amide bonds, likely due to the local cyclic groups atypical of peptide backbones. | ||
| Line 25: | Line 26: | ||
[[Image:Serine_protease_mechanism_by_snellios.png |thumb|left|Serine Protease Mechanism]] | [[Image:Serine_protease_mechanism_by_snellios.png |thumb|left|Serine Protease Mechanism]] | ||
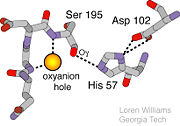
[[Image:Triad_1.jpg|thumb|left|Catalytic Triad]] | [[Image:Triad_1.jpg|thumb|left|Catalytic Triad]] | ||
| + | {{Clear}} | ||
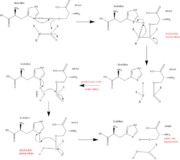
The function of Trypsin is to break down peptides using a hydrolysis reaction into amino acid building blocks. This mechanism is a general catalytic mechanism that all Serine proteases use. The active site where this mechanism occurs in Trypsin is composed of three amino acids and called a catalytic triad. The three catalytic residues are Serine 195, Histidine 57, and Aspartate 102 <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. The structure of the catalytic triad and the mechanism are shown in the figures to the right. In the mechanism, serine is bonded to the imidazole ring of the histidine. When histidine accepts a proton from serine an alkoxide nucleophile is formed. This nucleophile attacks the substrate when the substrate is present. The role of the aspartate residue is hold histidine in the proper position to make it a good proton acceptor. What makes this mechanism works is that a pocket if formed from the three residues and the three residues function to hold each other in proper position for nucleophilic attack. | The function of Trypsin is to break down peptides using a hydrolysis reaction into amino acid building blocks. This mechanism is a general catalytic mechanism that all Serine proteases use. The active site where this mechanism occurs in Trypsin is composed of three amino acids and called a catalytic triad. The three catalytic residues are Serine 195, Histidine 57, and Aspartate 102 <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. The structure of the catalytic triad and the mechanism are shown in the figures to the right. In the mechanism, serine is bonded to the imidazole ring of the histidine. When histidine accepts a proton from serine an alkoxide nucleophile is formed. This nucleophile attacks the substrate when the substrate is present. The role of the aspartate residue is hold histidine in the proper position to make it a good proton acceptor. What makes this mechanism works is that a pocket if formed from the three residues and the three residues function to hold each other in proper position for nucleophilic attack. | ||
The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. The mechanism can be followed in more detail in the figure on the right <ref>Image From: http://www.bmolchem.wisc.edu/courses/spring503/503-sec1/DRAWINGS/503-3a-2serineprotease.jpg</ref>. | The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. The mechanism can be followed in more detail in the figure on the right <ref>Image From: http://www.bmolchem.wisc.edu/courses/spring503/503-sec1/DRAWINGS/503-3a-2serineprotease.jpg</ref>. | ||
Revision as of 14:05, 23 December 2013
| |||||||||||
3D structures of Trypsin
Updated on 23-December-2013
Cationic trypsin
3nk8, 3nkk, 3mi4, 3mfj, 3iti, 2d8w, 2by5, 2by6, 2by7, 2by8, 2by9, 2bya, 2blv, 2blw, 2a7h, 1s0q, 1uto, 1utp, 1utq, 1utn, 1n6x, 1n6y, 1hj9, 2ptn, 3ptn, 5ptp, 3t25, 3t26, 3t27, 3t28, 3t29, 3unr, 4i8g, 4i8h, 4i8j, 4i8k, 4i8l - bTry1 - bovine
3qk1 – bTry1 (mutant)
1utk, 1utj, 1utl, 1utm, 1hj8 – Try1 – Salmon
1trn – hTry1 – human
3ljj, 3ljo, 3a7t, 3a7v, 3a7w, 3a7x, 3a7y, 3a7z, 3a80, 3a81, 3a82, 3a83, 3a84, 3a85, 3a86, 3a87, 3a88, 3a89, 3a8b, 3a8a, 3a8c, 3a8d, 3m35, 3aas, 3aau, 3aav, 3gy2, 3gy3, 3gy4, 3gy5, 3gy6, 3gy7, 3gy8, 2zq1, 2zq2, 2zhd, 2zfs, 2zft, 2zdk, 2zdl, 2zdm, 2zdn, 2oxs, 2otv, 2g8t, 2g5n, 2g5v, 2ah4, 2fx4, 2fx6, 1yp9, 2ayw, 1y3u, 1y3v, 1y3w, 1y3x, 1y3y, 1tx8, 1tx7, 1s0r, 1rxp, 1o2q, 1o2r, 1o2s, 1o2t, 1o2u, 1o2v, 1o2w, 1o2x, 1o2y, 1o2z, 1o30, 1o31, 1o32, 1o33, 1o34, 1o35, 1o36, 1o37, 1o38, 1o39, 1o3a, 1o3b, 1o3c, 1o3d, 1o3e, 1o3f, 1o3g, 1o3h, 1o3i, 1o3j, 1o3k, 1o3l, 1o3m, 1o3n, 1o3o, 1o3p, 1o2l, 1o2k, 1o2j, 1o2i, 1o2h, 1o2m, 1o2n, 1o2o, 1o2p, 1lqe, 1oyq, 1eb2, 1k1i, 1k1j, 1k1l, 1k1m, 1k1n, 1k1o, 1k1p, 1g36, 1j8a, 1jir, 1g3b, 1g3c, 1g3d, 1g3e, 1g9i, 1f0t, 1f0u, 1c1n, 1c1o, 1c1p, 1c1q, 1c1r, 1c1s, 1c1t, 1c2d, 1c2e, 1c2f, 1c2g, 1c2h, 1c2i, 1c2j, 1c2k, 1c2l, 1c2m, 1qbn, 1qbo, 1qb9, 1qb1, 1qb6, 1qa0, 1qcp, 1ce5, 2bza, 1az8, 1xuf, 1xug, 1bju, 1bjv, 1xuh, 1xui, 1xuj, 1xuk, 1auj, 2tio, 1tio, 1aq7, 3ati, 3atk, 3atl, 3atm, 3rxa, 3rxb, 3rxc, 3rxd, 3rxe, 3rxf, 3rxg, 3rxh, 3rxi, 3rxj, 3rxk, 3rxl, 3rxm, 3rxo, 3rxq, 3rxr, 3rxs, 3rxt, 3rxu, 3rxv - bTry1 + small molecule inhibitor
1v2j, 1v2l, 1v2m, 1v2n, 1v2o, 1v2p, 1v2q, 1v2r, 1v2s, 1v2t, 1v2u, 1v2v, 1v2w, 3plb, 3plk, 3plp, 3pm3, 3pmj, 3pwb, 3pwc, 3pyh, 3q00, 3unq, 3uns, 3uop, 3upe, 3uqo, 3uqv, 3uuz, 3uwi, 3uy9, 3v0x, 3v12, 3v13 - bTry1 (mutant) + small molecule inhibitor
3m7q, 2xtt, 3e8l, 3otj, 3i29, 3d65, 2qyi, 2qn5, 2o9q, 2plx, 2cmy, 2iln, 2uuy, 2j9n, 2g81, 2age, 2agg, 2agi, 2ftl, 2ftm, 2fi3, 2fi4, 2fi5, 1zr0, 1ox1, 1p2i, 1p2j, 1p2k, 1ejm, 1f2s, 3bte, 3btq, 3btd, 3btf, 3btg, 3bth, 3btk, 3btm, 3btt, 3btw, 2btc, 1sbw, 1taw, 1smf, 1ppc, 1ppe, 1pph, 2tld, 1tab, 1tpa, 1c9t, 1ezx, 2f3c, 3rdz – bTry1 + proteinase inhibitor
3ru4 – BTry1 + chymotrypsinogen
4b2b, 4b1t, 4b2a, 4b2c – bTry1 (mutant) + eglin (mutant)
2ra3, 1oph, 3veq - bTry1 (mutant) + proteinase inhibitor
1jrs, 1jrt, 1sfi, 1yyy, 1zzz, 4abi – bTry1 + polypeptide
1c5p, 1c5q, 1c5r, 1c5s, 1c5t, 1c5u, 1c5v, 1ghz, 1gi0, 1gi1, 1gi2, 1gi3, 1gi4, 1gi5, 1gi6, 1gj6, 1mts, 1mtu, 1mtv, 1mtw, 1ql7, 1ql8, 1ql9, 1v2k, 1y59, 1y5a, 1y5b, 1y5u, 3rxp, 4ab8, 4ab9, 4aba, 4abb, 4abd, 4abe, 4abf, 4abg, 4abh, 3vpk – bTry1 + inhibitor
4abj – bTry1 + Try inhibitor 1
4aoq, 4aor - bTry1 + Try inhibitor 3
4j2y - bTry1 + Try inhibitor
2eek – Try1 + inhibitor – Atlantic cod
Cationic trypsinogen
1tgc, 1tgt, 2tga, 2tgt, 1tgb, 1tld, 1tpo - bTryp1
1ntp - β-bTry1 – Neutron diffraction
1d6r, 4tpi, 1tgs, 2tgp, 3tpi, 2tpi, 2ptc - bTryp1 + proteinase inhibitor
1max, 1may, 1btp, 1bty, 1tps, 1tyn, 1tng, 1tnh, 1tni, 1tnj, 1tnk, 1tnl, 1gbt, 1tpp, 3ptb - bTry1 + small molecule inhibitor
1btw, 1btx, 1btz - bTry1 + polypeptide
Anionic trypsin
2zpq, 2zpr, 2zps, 1mbq – Try2 – Chum salmon
1bit, 2tbs - AsTry2 – Atlantic salmon
2sta, 2stb, 1bzx - AsTry2 + proteinase inhibitor
1a0j - AsTry2 + small molecule inhibitor
1ane, 1bra - rTry2]] - rat
1amh, 1dpo, 1anb, 1anc, 1and, 1trm, 2trm - rTry2 (mutant)
3fp6, 3tgi, 1brb, 1brc – rTry2 + proteinase inhibitor
3fp7, 3fp8, 1ykt, 1ylc, 1yld, 1co7, 1k9o, 1slu, 1slv, 1slw, 1slx - rTry2 (mutant) + proteinase inhibitor
1j14, 1j15, 1j16, 1j17 - rTry2 (mutant) + small molecule inhibitor
Anionic trypsinogen
1f5r, 1f7z, 3tgk, 1ezs, 1ezu, 3tgj - rTryp2 (mutant) + proteinase inhibitor
1fy8 - rTryp2 + proteinase inhibitor
Trypsinogen
1tgn – bTryp
2tgd – bTryp + inhibitor
Mesotrypsin
3l33 – hTry3 (mutant) + amyloid β A4
3l3t - hTry3 residues 28-251 (mutant) + amyloid β precursor
2r9p – hTry3 (mutant) + BPTI
Brain trypsin
1h4w – hTry4 + small molecule inhibitor
Neurotrypsin
2k4r, 2k51 – rNTry Kringle domain – NMR
Streptomyces griseus trypsin
3i77, 3i78, 1os8, 1sgt – SGT – Streptomyces griseus
3beu, 2fmj – SGT (mutant)
1oss - SGT (mutant) + small molecule inhibitor
1s81 – pTry – pig
1aks - α-pTry
1ept - ε-pTry
1mct - β-pTry + proteinase inhibitor
3myw, 1yf4, 1z7k, 1tx6, 1v6d, 1uhb, 1h9h, 1h9i, 1eja, 1c9p, 1avw, 1avx, 1ldt, 1tfx, 1an1, 4an7 – pTry + proteinase inhibitor
2a31, 2a32, 1s5s, 1s6f, 1s6h, 1s82, 1s83, 1s84, 1s85, 1fmg, 1fn6, 1fni, 1qqu – pTry + small molecule inhibitor
2vu8 – Try + proteinase inhibitor – mold
2g51, 2g52, 2g55, 1xvo, 1pq5, 1pq7 – FoTry – Fusarium oxysporum
1ppz, 1pqa, 1try - FoTry + small molecule inhibitor
1xvm, 1pq8, 1fn8, 1fy4, 1fy5, 1gdn, 1gdq, 1gdu – FoTry + polypeptide
2f91 – Try-hepatopancreas - Crayfish
References
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.
- ↑ Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Image From:

- ↑ Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Joshi RS, Mishra M, Tamhane VA, Ghosh A, Sonavane U, Suresh CG, Joshi R, Gupta VS, Giri AP. The remarkable efficiency of a Pin-II proteinase inhibitor sans two conserved disulfide bonds is due to enhanced flexibility and hydrogen bond density in the reactive site loop. J Biomol Struct Dyn. 2012 Dec 20. PMID:23256852 doi:10.1080/07391102.2012.745378
- ↑ 12.0 12.1 Green TR, Ryan CA. Wound-Induced Proteinase Inhibitor in Plant Leaves: A Possible Defense Mechanism against Insects. Science. 1972 Feb 18;175(4023):776-7. PMID:17836138 doi:10.1126/science.175.4023.776
- ↑ Kong L, Ranganathan S. Tandem duplication, circular permutation, molecular adaptation: how Solanaceae resist pests via inhibitors. BMC Bioinformatics. 2008;9 Suppl 1:S22. PMID:18315854 doi:10.1186/1471-2105-9-S1-S22
- ↑ Johnson R, Narvaez J, An G, Ryan C. Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9871-5. PMID:2602379
- ↑ Duan X, Li X, Xue Q, Abo-el-Saad M, Xu D, Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol. 1996 Apr;14(4):494-8. PMID:9630927 doi:10.1038/nbt0496-494
- ↑ Nielsen KJ, Heath RL, Anderson MA, Craik DJ. Structures of a series of 6-kDa trypsin inhibitors isolated from the stigma of Nicotiana alata. Biochemistry. 1995 Nov 7;34(44):14304-11. PMID:7578034
- ↑ Scanlon MJ, Lee MC, Anderson MA, Craik DJ. Structure of a putative ancestral protein encoded by a single sequence repeat from a multidomain proteinase inhibitor gene from Nicotiana alata. Structure. 1999 Jul 15;7(7):793-802. PMID:10425681
- ↑ Lee MC, Scanlon MJ, Craik DJ, Anderson MA. A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nat Struct Biol. 1999 Jun;6(6):526-30. PMID:10360353 doi:10.1038/9293
- ↑ Schirra HJ, Scanlon MJ, Lee MC, Anderson MA, Craik DJ. The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata. J Mol Biol. 2001 Feb 9;306(1):69-79. PMID:11178894 doi:10.1006/jmbi.2000.4318
- ↑ Schirra HJ, Craik DJ. Structure and folding of potato type II proteinase inhibitors: circular permutation and intramolecular domain swapping. Protein Pept Lett. 2005 Jul;12(5):421-31. PMID:16029154
- ↑ Schirra HJ, Anderson MA, Craik DJ. Structural refinement of insecticidal plant proteinase inhibitors from Nicotiana alata. Protein Pept Lett. 2008;15(9):903-9. PMID:18991765
- ↑ Schirra HJ, Guarino RF, Anderson MA, Craik DJ. Selective removal of individual disulfide bonds within a potato type II serine proteinase inhibitor from Nicotiana alata reveals differential stabilization of the reactive-site loop. J Mol Biol. 2010 Jan 22;395(3):609-26. Epub 2009 Nov 17. PMID:19925809 doi:10.1016/j.jmb.2009.11.031
- ↑ Li XQ, Zhang T, Donnelly D. Selective loss of cysteine residues and disulphide bonds in a potato proteinase inhibitor II family. PLoS One. 2011 Apr 11;6(4):e18615. PMID:21494600 doi:10.1371/journal.pone.0018615
- ↑ Barrette-Ng IH, Ng KK, Cherney MM, Pearce G, Ryan CA, James MN. Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg. J Biol Chem. 2003 Jun 27;278(26):24062-71. Epub 2003 Apr 8. PMID:12684499 doi:10.1074/jbc.M302020200
- ↑ Dunse KM, Kaas Q, Guarino RF, Barton PA, Craik DJ, Anderson MA. Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15016-21. Epub 2010 Aug 9. PMID:20696921 doi:10.1073/pnas.1009327107
- ↑ Tamhane VA, Giri AP, Kumar P, Gupta VS. Spatial and temporal expression patterns of diverse Pin-II proteinase inhibitor genes in Capsicum annuum Linn. Gene. 2009 Aug 1;442(1-2):88-98. Epub 2009 Apr 22. PMID:19393726 doi:10.1016/j.gene.2009.04.012
- ↑ Tamhane VA, Chougule NP, Giri AP, Dixit AR, Sainani MN, Gupta VS. In vivo and in vitro effect of Capsicum annum proteinase inhibitors on Helicoverpa armigera gut proteinases. Biochim Biophys Acta. 2005 Mar 11;1722(2):156-67. Epub 2005 Jan 12. PMID:15715970 doi:10.1016/j.bbagen.2004.12.017
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, Leah Bowlin, David Canner, Glenn Jones, Ben Hallowell, Karl Oberholser, Jaime Prilusky