Sandbox Reserved 770
From Proteopedia
(→Structure) |
(→Structure) |
||
| Line 47: | Line 47: | ||
[[Image:Tertiary Structure.gif|thumb|left|300px|Figure 4. Tertiary structure of PAL. The three central core helices, leading to the active site, are colored blue, green, and yellow. (a) Stereoview of the PAL monomer with residue numbering. (b) Stereoview of MIO and Phe413 interactions with the three central helices, polarized with their N termini directed toward the active site. Hydrogen bonds are indicated by dashed lines. <ref name=crystal>http://pubs.acs.org.prox.lib.ncsu.edu/doi/pdfplus/10.1021/bi049053%2B</ref>]] | [[Image:Tertiary Structure.gif|thumb|left|300px|Figure 4. Tertiary structure of PAL. The three central core helices, leading to the active site, are colored blue, green, and yellow. (a) Stereoview of the PAL monomer with residue numbering. (b) Stereoview of MIO and Phe413 interactions with the three central helices, polarized with their N termini directed toward the active site. Hydrogen bonds are indicated by dashed lines. <ref name=crystal>http://pubs.acs.org.prox.lib.ncsu.edu/doi/pdfplus/10.1021/bi049053%2B</ref>]] | ||
| - | The central core within each monomer of PAL contains three central alpha helices of triple-coiled coils, improving its rigidity similar to fibrous proteins in keratin. The three core helices are oriented with similarly aligned dipoles to create an electro-positive platform for cofactor MIO to anchor through noncovalent bonding, shown in Figure 4b. Covalent linkages between cofactor MIO and PAL backbone act to direct the C-terminus of MIO to an alpha helix's N-terminus, creating a loop, thus the helix's positive pole points toward the cofactor and the active site. | + | The central core within each monomer of PAL contains three central alpha helices of triple-coiled coils (Helix 3,Helix 4, and Helix 7), improving its rigidity similar to fibrous proteins in keratin. The three core helices are oriented with similarly aligned dipoles to create an electro-positive platform for cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO) to anchor through noncovalent bonding, shown in Figure 4b. Covalent linkages between cofactor MIO and PAL backbone act to direct the C-terminus of MIO to an alpha helix's N-terminus, creating a loop, thus the helix's positive pole points toward the cofactor and the active site. PAL active site contains a highly conserved Ala-Ser-Gly triad. Post-translational modification of an electrophilic prostetic group MIO formed autocatalytically by cyclization and dehydration of PAL's active site. MIO provides the surface for phenylalanine conversion to produce trans-cinnamic acid and ammonia. |
| + | |||
| + | Most of the conserved active site residues are contained within seven stabilizing alpha helices. Six positive alpha helices point toward the active site in association with MIO cofactor. This association will not only increase the electrophilicity of MIO, but also increases he positive charge of Lys residue 468. The positive poles stabilize the carbanion intermediate's negative charge introduced by the elimination unimolecular conjugate base (E1cB) mechanism of substrate phenylalanine, promoting the interactions between the negatively charged carboxylate end of the substrate. Promoting this interaction will subsequently stabilize the carbanion intermediate as well. | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Revision as of 19:49, 6 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Phenylalanine Ammonia Lyase
|
Phenylalanine Ammonia Lyase
Phenylalanine Ammonia Lyase (PAL) catalyses the first and committed step in the phenyl propanoid pathway, forming trans-cinnamic acid by non-oxidative deamination of L-phenylalanine. Biosynthesis of polyphenol compounds are found in lignin in plants, some yeast and fungi. [1]
PAL is the most studied enzyme corcerning secondary metabolism in plants due to the significant fluctuations in enzyme levels within relatively short time intervals in response to a variety of stimuli. Activation of PAL is induced in response to various stimuli such as tissue wounding, pathogenic attack, ultra-violet light radiation, low temperatures, and hormones. PAL substitution therapy has been used in phenylketonuria treatment. The reverse reaction of PAL is used to create L-phenylalanine, a precursor for Aspartame sweetener. [2]
Contents |
General Information
Gene Name: PAL [3]
Organism: Rhodosporidium toruloides [3]
Classification: Lyase [4]
Sequence Length: 1432 Residues [5]
Molecular Weight: 155505.58 Da [6]
Isoelectric Point: 6.68 [5]
Chains: A, B [6]
Ligands: 715 [7], 4-Carboxycinnamic Acid (CIN) [8], Selenomethionine (MSE) [9]
Kinetic Parameters: Km=0.29 mM [3]
pH Dependence: Optimal pH=8.5 [3]
Structure


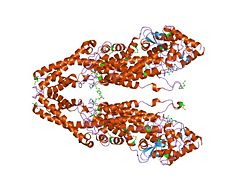
Phenylalanine ammonia lyase enzyme is a dimer composed of two identical subunits. [11] Each subunit of PAL from R. toruloides assume a "seahorse" shape by interlocking head-to-tail, creating overlapping regions with two adjacent subunits, as shown by Figure 2. These overlapping regions maximize interactions between subunits, giving rise to the formation of the tightly assembled tetramer, as shown in Figure 3. Formation of the tetramer buries 58% of their combined surfaces. [12] Of the 66 interactions between adjacent subunits, 25 hydrogen bonding interactions exists between Asp & Glu carboxylate oxygens and NH2 & OH moieties, including a prominent band of Asp & Glu interactions with Arg side chains between subunits nearby the central bundle of helices. PAL's central core is comprised of parallel alpha helices of varying lengths. There is only one section of Beta sheet longer than three residues in PAL, which resides in the funnel region leading to the active site. PAL and HAL (Histadine Ammonia Lyase) contain similar folds, but PAL differs from HAL with 215 additional residues. Of the 215 residues from this section, 155 residues extend above and below the main body of the structure, creating a "fan" arrangement, shown as the bracketed areas in Figure 3a.

The central core within each monomer of PAL contains three central alpha helices of triple-coiled coils (Helix 3,Helix 4, and Helix 7), improving its rigidity similar to fibrous proteins in keratin. The three core helices are oriented with similarly aligned dipoles to create an electro-positive platform for cofactor 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO) to anchor through noncovalent bonding, shown in Figure 4b. Covalent linkages between cofactor MIO and PAL backbone act to direct the C-terminus of MIO to an alpha helix's N-terminus, creating a loop, thus the helix's positive pole points toward the cofactor and the active site. PAL active site contains a highly conserved Ala-Ser-Gly triad. Post-translational modification of an electrophilic prostetic group MIO formed autocatalytically by cyclization and dehydration of PAL's active site. MIO provides the surface for phenylalanine conversion to produce trans-cinnamic acid and ammonia.
Most of the conserved active site residues are contained within seven stabilizing alpha helices. Six positive alpha helices point toward the active site in association with MIO cofactor. This association will not only increase the electrophilicity of MIO, but also increases he positive charge of Lys residue 468. The positive poles stabilize the carbanion intermediate's negative charge introduced by the elimination unimolecular conjugate base (E1cB) mechanism of substrate phenylalanine, promoting the interactions between the negatively charged carboxylate end of the substrate. Promoting this interaction will subsequently stabilize the carbanion intermediate as well.
References
- ↑ http://www.sciencedirect.com.prox.lib.ncsu.edu/science/article/pii/S0031942200804653
- ↑ http://www.sciencedirect.com.prox.lib.ncsu.edu/science/article/pii/0031942273850010#
- ↑ 3.0 3.1 3.2 3.3 http://www.uniprot.org/uniprot/P11544
- ↑ http://oca.weizmann.ac.il/oca-bin/ocashort?id=1T6J
- ↑ 5.0 5.1 http://isoelectric.ovh.org/files/calculate.php
- ↑ 6.0 6.1 http://www.rcsb.org/pdb/explore/explore.do?structureId=1T6J
- ↑ http://oca.weizmann.ac.il/oca-bin/send-het?175
- ↑ http://oca.weizmann.ac.il/oca-bin/send-het?CIN
- ↑ http://oca.weizmann.ac.il/oca-bin/send-het?MSE
- ↑ http://maptest.rutgers.edu/drupal/?q=node/408
- ↑ http://ci.nii.ac.jp/els/110006324658.pdf?id=ART0008332067&type=pdf&lang=en&host=cinii&order_no=&ppv_type=0&lang_sw=&no=1386328093&cp=
- ↑ 12.0 12.1 http://pubs.acs.org.prox.lib.ncsu.edu/doi/pdfplus/10.1021/bi049053%2B