Chymotrypsin
From Proteopedia
(→Substrate binding and catalysis) |
|||
| Line 1: | Line 1: | ||
[[Image:2ea3.png|left|200px|thumb|Crystal Structure of ''Cellulomonas Bogoriensis'' Chymotrypsin [[2ea3]]]] | [[Image:2ea3.png|left|200px|thumb|Crystal Structure of ''Cellulomonas Bogoriensis'' Chymotrypsin [[2ea3]]]] | ||
| - | + | [[Chymotrypsin]] (Chy or α-Chy) is a digestive enzyme containing an active serine residue, which helps to digest proteins in our food. Other related proteases are crucial for blood clotting ([http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer&part=A1378&rendertype=figure&id=A1401 thrombin and other proteases]), for the AIDS virus metabolism ([http://www.proteopedia.org/wiki/index.php/Hiv_protease HIV protease]) and for many other processes relevant to human health and agriculture. Chymotrypsin cleaves peptide bonds of proteins where the amide side of the bond is an aromatic amino acid like tyrosine, phenylalanine or tryptophan. The image at the left is the crystal structure of chymotrypsin from ''Cellulomonas Bogoriensis'' ([[2ea3]]) with sulfate ions. Below is description of the structure of bovine chymotrypsin. Some additional details in<br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Chymotrypsin]] (Chy or α-Chy) is a digestive enzyme containing an active serine residue. | + | |
| - | + | ||
*[[Molecular Playground/Chymotrypsin]]<br /> | *[[Molecular Playground/Chymotrypsin]]<br /> | ||
*[[Serine Proteases]]. | *[[Serine Proteases]]. | ||
{{TOC limit|limit=2}} | {{TOC limit|limit=2}} | ||
| - | |||
| - | |||
| - | {{STRUCTURE_1gl0 | PDB=1glo | SCENE='Chymotrypsin/Chymotrypsin_triad/2' }} | ||
== Overview == | == Overview == | ||
| - | + | <Structure load='7gch' size='300' frame='true' align='right' caption='[[7gch]] Bovine chymotrypsin with bound inhibitor' scene='38/387136/Bovine_chymotrypsin_overview/1' />While chymotrypsin occurs in many organisms, the most-studied chymotrypsin is that from cows (bovine chymotrypsin), shown here with an inhibitor molecule bound to the active site (<scene name='38/387136/Bovine_chymotrypsin_overview/1'>default scene</scene>). It is synthesized as a single polypeptide chain of 245 amino acids, called chymotrypsinogen, that is inactive. The enzyme is activated by one cleavage by trypsin and two cleavages by chymotrypsin (autolytic cleavages) that result in the loss of four amino acids from the remaining three polypeptides, shown here in turquoise, beige, and violet. These three chains are held together by <scene name='38/387136/Bovine_chymotrypsin_overview/5'> two inter-chain disulfide bonds</scene>. The bonded cysteine residues are shown in space fill with yellow sulfur atoms. There also three <scene name='38/387136/Bovine_chymotrypsin_overview/6'>intra-chain disulfide bonds</scene>. Here chymotrypsin is shown in cartoon with pink α-helices and yellow β-strands, and this shows that it is mainly composed of <scene name='38/387136/Bovine_chymotrypsin_overview/7'>two beta barrels</scene>. | |
| - | + | ||
| - | + | == Substrate-binding and Active Sites == | |
| - | + | [[Image:LPFstructure.jpg|left]]Features of the substrate-binding and active sites can be seen in the structure of bovine chymotrypsin bound to the inhibitor N-acetyl-L-leucyl-L-phenylalanyl trifuoromethyl ketone, which resembles a peptide substrate (see structure in left figure). The colored backgrounds in the figure indicate the four components of structure and shows the bond (yellow on black background) that is position to be cleaved. | |
| - | = | + | Here is chymotrypsin (space fill) with the inhibitor (CPK ball & stick) showing the <scene name='38/387136/Bovine_chymotrypsin_overview/3'>inhibitor sitting in the active site</scene>. Note the active site is in a depression on the surface of the enzyme. Chymotrypsin contains three residues, Ser 195, His 57 and Asp 102, which are known as its <scene name='38/387136/Bovine_chymotrypsin_active_sit/4'>catalytic triad</scene>, shown in CPK ball and stick in this close up of the active site. Similar three-dimensional arrangements of a serine, a histidine and an aspartate are observed in many other proteases, and the role of these three residues in catalysis has been studied extensively. Serine acts as a nucleophile (contributing the electron pair for a new bond) attacking the carbonyl carbon of the peptide bond to be hydrolyzed. Histidine and aspartate turn serine into a better nucleophile by assisting in removing a hydrogen ion from serine. |
| - | + | ||
| - | + | The <scene name='38/387136/Bovine_chymotrypsin_active_sit/1'>substrate-binding site</scene>, can be seen in this view with the inhibitor in light gray ball & stick with its phenyl group in orchid. By moving the structure back and forth with your mouse, it is easy to see that the phenyl group is located in the hydrophobic binding pocket of the enzyme. This binding pocket determines the enzyme's preference for cleavage of peptides on the C-terminal side of aromatic residues. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | This view shows the | + | This view shows the <scene name='38/387136/Bovine_chymotrypsin_active_sit/3'>carbonyl group of the inhibitor</scene> in CPK colors. The triflouromethyl group is bound to the carbonyl carbon via the yellow bond. In a peptide substrate, the triflouromethyl group would be replaced by the first amino acid residue of the rest of the peptide chain, and the yellow bond would be the bond that is cleaved. The carbonyl carbon of the inhibitor is 1.95 Å away from the side chain oxygen of serine 195, and this indicates they are covalently bound (bond indicated by dotted line). Thus, this structure is similar to the '''tetrahedral intermediate''' that is formed during the cleavage reaction. The negative charge that develops on the carbonyl oxygen of the substrate is stabilized by hydrogen bonds to the backbone nitrogens of Ser 195 and Gly 193, shown in blue spacefill. The hydrogen atoms involved in these hydrogen bonds are not shown. |
== 3D Structures of Chymotrypsin == | == 3D Structures of Chymotrypsin == | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | The Chy precursor is the inactive '''chymotrypsinogen''' (Chygen) which gets cleaved 3 times by trypsin and chymotrypsin losing a 4 amino acid long peptide to become the active Chy. '''γ-Chy''' is a covalent acyl adduct of '''α-Chy'''. '''δ-Chy''' results when Chygen is cleaved only twice. | ||
=== Native Chymotrypsin === | === Native Chymotrypsin === | ||
Revision as of 17:26, 23 December 2013
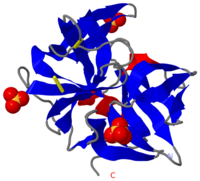
Chymotrypsin (Chy or α-Chy) is a digestive enzyme containing an active serine residue, which helps to digest proteins in our food. Other related proteases are crucial for blood clotting (thrombin and other proteases), for the AIDS virus metabolism (HIV protease) and for many other processes relevant to human health and agriculture. Chymotrypsin cleaves peptide bonds of proteins where the amide side of the bond is an aromatic amino acid like tyrosine, phenylalanine or tryptophan. The image at the left is the crystal structure of chymotrypsin from Cellulomonas Bogoriensis (2ea3) with sulfate ions. Below is description of the structure of bovine chymotrypsin. Some additional details in
Contents |
Overview
|
Substrate-binding and Active Sites
Features of the substrate-binding and active sites can be seen in the structure of bovine chymotrypsin bound to the inhibitor N-acetyl-L-leucyl-L-phenylalanyl trifuoromethyl ketone, which resembles a peptide substrate (see structure in left figure). The colored backgrounds in the figure indicate the four components of structure and shows the bond (yellow on black background) that is position to be cleaved.Here is chymotrypsin (space fill) with the inhibitor (CPK ball & stick) showing the . Note the active site is in a depression on the surface of the enzyme. Chymotrypsin contains three residues, Ser 195, His 57 and Asp 102, which are known as its , shown in CPK ball and stick in this close up of the active site. Similar three-dimensional arrangements of a serine, a histidine and an aspartate are observed in many other proteases, and the role of these three residues in catalysis has been studied extensively. Serine acts as a nucleophile (contributing the electron pair for a new bond) attacking the carbonyl carbon of the peptide bond to be hydrolyzed. Histidine and aspartate turn serine into a better nucleophile by assisting in removing a hydrogen ion from serine.
The , can be seen in this view with the inhibitor in light gray ball & stick with its phenyl group in orchid. By moving the structure back and forth with your mouse, it is easy to see that the phenyl group is located in the hydrophobic binding pocket of the enzyme. This binding pocket determines the enzyme's preference for cleavage of peptides on the C-terminal side of aromatic residues.
This view shows the in CPK colors. The triflouromethyl group is bound to the carbonyl carbon via the yellow bond. In a peptide substrate, the triflouromethyl group would be replaced by the first amino acid residue of the rest of the peptide chain, and the yellow bond would be the bond that is cleaved. The carbonyl carbon of the inhibitor is 1.95 Å away from the side chain oxygen of serine 195, and this indicates they are covalently bound (bond indicated by dotted line). Thus, this structure is similar to the tetrahedral intermediate that is formed during the cleavage reaction. The negative charge that develops on the carbonyl oxygen of the substrate is stabilized by hydrogen bonds to the backbone nitrogens of Ser 195 and Gly 193, shown in blue spacefill. The hydrogen atoms involved in these hydrogen bonds are not shown.
3D Structures of Chymotrypsin
Updated on 23-December-2013
The Chy precursor is the inactive chymotrypsinogen (Chygen) which gets cleaved 3 times by trypsin and chymotrypsin losing a 4 amino acid long peptide to become the active Chy. γ-Chy is a covalent acyl adduct of α-Chy. δ-Chy results when Chygen is cleaved only twice.
Native Chymotrypsin
1yph – bChyA chain A - bovine
4cha, 5cha – BtChy
1kdq – rChyB, chain B (mutant) - rat
2ea3 – Chy – Cellulomonas bogoriensis
1ab9, 8gch, 1gct, 2gct, 3gct, 2gch - gamma BtChy
Native Chymotrypsinogen
2cga, 1chg – bChygen A
2jet – rChygen B chain A,B
Chymotrypsin + polypeptide inhibitors
1cbw, 1mtn - bChy+BPTI
1t7c, 1t8l, 1t8m, 1t8n, 1t8o – bChyA+P1 BPTI variants
1oxg – bChyA+autolysis peptide
1p2m, 1p2n, 1p2o, 1p2q – bChyA+ 4 amino acids in S1 pocket
1n8o – bChyA+ecotin
1ca0 – bChy+APPI
1acb, 4h4f – bChy+Elgin C
2cho – bChy+turkey ovomucoid third domain
Chymotrypsin + inhibitors
3bg4 – ChyA chain A+guamerin
2p8o - bChyA chain A+benzohydroxamic acid/vanadate
1eq9 – Chy+PMSF – fire ant
2cha – bChy+p-sulfinotoluene
γ-Chymotrypsin + inhibitors
1gg6 – γ-bChy+N-acetyl-phenylalanine trifluoromethyl ketone
1ggd – γ-bChy+N-acetyl-phenylalanine trifluoromethyl aldehyde
1afq - γ-bChy+synthetic inhibitor
3gch, 4gch, 5gch - γ-bChy+cinnamate
6gch, 7gch - γ-bChy+trifluoromethy ketone
6cha – γ-bChy+phenylethane boronic acid – transition state inhibitor
1gmc, 1gmd – γ-bChy+hexane – transition state inhibitor
2gmt - γ-bChy+N-acetyl-alanyl-phenylalanyl-chloroethyl ketone
1gmh, 1gcd - γ-bChy+organophosphoryl
1gha, 1ghb - γ-bChy+ N-acetyl-tryptophan
γ-Chymotrypsin + reaction transition state inhibitors
6cha – γ-bChy+phenylethane boronic acid
1gmc, 1gmd – γ-bChy+hexane
δ-Chymotrypsin + inhibitors
1dlk – δ-bChy+peptidyl chloromethyl ketone
Chymotrypsinogen + inhibitors
1gl0, 1gli – ChygenA+PMP_D2v – Locusta migratoria
1k2i - bChygen+7-hydroxycoumarin
2y6t – bChygenA + ecotin
3t62 - bChygenA + Kunitz-type proteinase inhibitor SHPI-1
Further reading
You can learn more about chymotrypsin structure, function and regulation in this publicly available chapter of the Biochemistry textbook by Berg, Tymoczka and Stryer.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alice Harmon, Alexander Berchansky

