Amylase
From Proteopedia
| Line 1: | Line 1: | ||
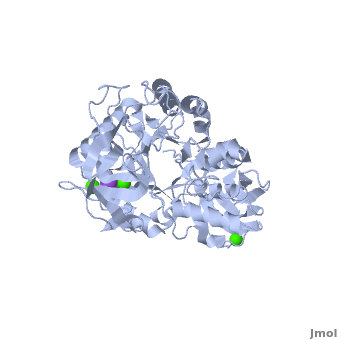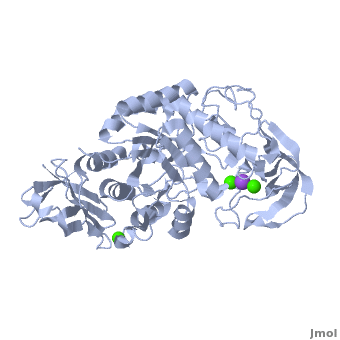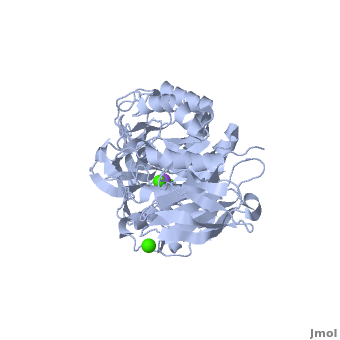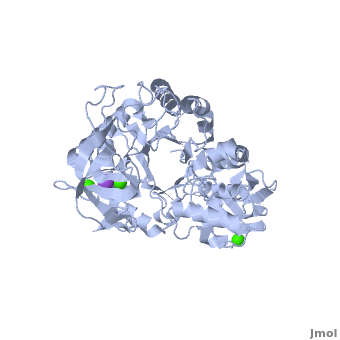
| - | <StructureSection load='1hvx' size='400' side='right' scene='Sandbox_182/Alpha-amylase/1' caption=''> | + | <StructureSection load='1hvx' size='400' side='right' scene='Sandbox_182/Alpha-amylase/1' caption='Amylase complex with Ca+2 and Na+ ions (PDB code [[1hvx]])'> |
=Introduction= | =Introduction= | ||
Discovered and isolated by [http://en.wikipedia.org/wiki/Anselme_Payen Anselme Payen] in 1833, amylase was the first enzyme to be discovered<ref name="book">Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press</ref>. Amylases are hydrolases, acting on α-1,4-glycosidic bonds<ref name="Path">PMID:9541387</ref>. They can be further subdivided into α,β and γ amylases<ref name="book"/>.'''α-Amylase''' (AAM) is an enzyme that acts as a catalyst for the hydrolysis of alpha-linked polysaccharides into α-anomeric products<ref name="Main">PMID:11226887</ref>. The enzyme can be derived from a variety of sources, each with different characteristics. α-Amylase found within the human body serves as the enzyme active in pancreatic juice and salvia<ref name="Path"/>. α-Amylase is not only essential in human physiology but has a number of important biotechnological functions in various processing industries. Beta/alpha amylase (BAAM) is a precursor protein which is cleaved to form the beta-amylase and alpha-amylase after secretion. | Discovered and isolated by [http://en.wikipedia.org/wiki/Anselme_Payen Anselme Payen] in 1833, amylase was the first enzyme to be discovered<ref name="book">Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press</ref>. Amylases are hydrolases, acting on α-1,4-glycosidic bonds<ref name="Path">PMID:9541387</ref>. They can be further subdivided into α,β and γ amylases<ref name="book"/>.'''α-Amylase''' (AAM) is an enzyme that acts as a catalyst for the hydrolysis of alpha-linked polysaccharides into α-anomeric products<ref name="Main">PMID:11226887</ref>. The enzyme can be derived from a variety of sources, each with different characteristics. α-Amylase found within the human body serves as the enzyme active in pancreatic juice and salvia<ref name="Path"/>. α-Amylase is not only essential in human physiology but has a number of important biotechnological functions in various processing industries. Beta/alpha amylase (BAAM) is a precursor protein which is cleaved to form the beta-amylase and alpha-amylase after secretion. | ||
Revision as of 12:02, 20 January 2014
| |||||||||||
3D structures of Amylase
Updated on 20-January-2014
α-amylase
3ij7, 1xv8, 1c8q, 1bsi, 1smd, 1hny – hAAM – human
1xgz, 1q4n, 1kb3, 1kbb, 1kbk, 1kgu, 1kgw, 1kgx, 1jxj, 1jxk, 2cpu - hAAM (mutant)
3n8t – TkAAM – Thermococcus kodakarensis
3k8k – BtAAM – Bacterioides thetaiotaomicron
3kwx, 2guy – AoAAM – Aspergilluys oryzae
2wpg – AAM – Xanthomonas campestris
2wc7, 2wcs, 2wkg – AAM catalytic region – Cyanobacterium
3bh4 – BaAAM – Bacillus amyloliquefaciens
3bsg – bAAM (mutant)
3dhu – AAM – Lactobacillus plantarum
3dc0 – AAM – Bacillus KR8104
3bcf, 1wza – HoAAM – Halothermothrix orenii
2die, 1wp6 – AAM alkaline – Bacillus sp.
1ud2, 1ud4, 1ud5, 1ud6, 1ud8 - AAM – Bacillus sp. KSM-K38
1ud3 – AAM (mutant) – Bacillus sp. KSM-K38
2gjr – BhAAM – Bacillus halmapalus
2b5d – AAM – Thermotoga maritima
2c3g, 2c3v – BhaloAAM – Bacillus halodurans
1ji1, 1ji2, 1bvz - TvAAM – Thermoactinomyces vulgaris
1wzk, 1wzl, 1wzm, 1izj, 1izk, 1jf5, 1jf6 – TvAAM (mutant)
1mwo, 1mxd – PwAAM – Pyrococcus woesei
1ob0, 1bli - BlAAM (mutant) – Bacillus licheniformis
1vjs – BlAAM precursor
1bpl – BlAAM
1b0i, 1aqm, 1aqh – PhAAM - Pseudoalteromonas haloplanktis
1g5a – NpAAM – Neisseria polysaccharea
1hvx - BaAAM – Bacillus stearothermophilus
1qho – GsAAM – Geobacillus stearothermophilus
1jae – TmAAM – Tenebrio molitor
1pif – pAAM – pig
6taa, 2aaa - AoAAM
4aee – AAM catalytic domain – Staphylothermus marinus
3ren – AAM – Clostridium perfringens
3vm5 – AAM – Oryzias latipes
3vm7 – AAM – Malbranchea cinnamomea
4gkl – AAM – Thermotoga neapolitana
AAM binary complexes
1xd0, 1xd1, 1cpu, 1jfh - hAAM + saccharide
1b2y, 1xcw, 1xcx - hAAM + acarbose
3blk, 3blp, 1z32, 1nm9, 1mfu, 1mfv, 3cpu - hAAM (mutant) + saccharide
3dhp, 1xh0, 1xh2 - hAAM (mutant) + acarbose
3ij8, 3ij9 – hAAM catalytic intermediate
3baw – hAAM + N3
3bax - hAAM (mutant) + N3
3bak – hAAM (mutant) + NO3
1xh1 - hAAM (mutant) + Cl
2qv4, 3baj, 3bay - hAAM + acarbose + NO2
3old, 3ole, 3olg, 3oli – hAAM + statin
1u2y, 1u30, 1u33 – hAAM + inhibitor
4gqq – hAAM + ethyl caffeate
4gqr – hAAM + myricetin
3n92, 3n98 – TkAAM + saccharide
3l2l, 3l2m, 1vah, 1wo2, 1ua3, 1pig, 1ppi - pAAM + saccharide
1hx0 – pAAM + acarbose
1kxq, 1kxt, 1kxv – pAAM + antibody VHH fragment
1bvn, 1dhk – pAAM + protein inhibitor
1ose – pAAM + acarbose
3k8l - BtAAM (mutant) + saccharide
3k8m - BtAAM + acarbose
2d2o, 1jl8, 1jib - TvAAM + saccharide
3a6o – TvAAM + acarbose
2d0f, 2d0g, 2d0h, 1vb9, 1vfm, 1vfo, 1vfu, 1uh2, 1uh4 - TvAAM (mutant) + saccharide
1uh3 - TvAAM (mutant) + acarbose
1ava – bAAM + protein inhibitor
1bg9, 1rpk - bAAM + acarbose
1p6w – bAAM + substrate analog
1rp8, 1b1y, 1rp9 - bAAM (mutant) + saccharide
3bsh, 2qpu, 2qps - bAAM (mutant) + acarbose
3bcd - HoAAM + saccharide
3bc9 - HoAAM + acarbose
2gjp, 1w9x - BhAAM + saccharide
2gvy - AoAAM + saccharide
1zs2, 1s46, 1mvy, 1mw0, 1mw1, 1mw2, 1mw3 - NpAAM (mutant) + saccharide
1bag - BsAAM + saccharide – Bacillus subtilis
1ua7 – BsAAM + acarbose
2d3l, 2d3n - BacAAM + saccharide – Bacillus
2c3h, 2c3w, 2c3x - BhaloAAM + saccharide
1mxg - PwAAM + acarbose
1g9h, 1g94 - PhAAM + saccharide
1kxh - PhAAM (mutant) + acarbose
1l0p – PhAAM + NO3
1e40 – BaAAM chimera + saccharide
1e3z - BaAAM chimera + acarbose
1fa2 - AAM + saccharide – Sweet potato
1qhp - GsAAM + saccharide
4e2o - GsAAM + acarbose
1clv, 1viw, 1tmq – TmAAM + protein inhibitor
1gah, 1gai – AaAAM + acarbose – Aspergillus awamori
3gly, 1agm, 1glm – AaAAM + saccharide
Pullulanase α-amylase
2wan – AAM – Bacillus acidopullululyticus
2fgz – KaAAM – Klebsiella aerogenes
2e8y - BsAAM
1ji2 - TvAAM
1jl5, 1jf6, 1wzk, 1wzl, 1wzm - TvAAM (mutant)
Pullulanase α-amylase binary complexes
2fh6, 2fh8, 2fhb, 2fhc, 2fhf - KaAAM + saccharide
3fax - AAM + saccharide – Streptococcus agalactiae
2e8z, 2e9b - BsAAM + saccharide
2d2o - TvAAM + saccharide
1g1y, 1jib, 1jl8, 1vfm, 1vfo, 1vfu, 1vb9 - TvAAM (mutant) + saccharide
3a6o – TvAAM + acarbose
Neopullulanase α-amylase
4aef – AAM – Pyrococcus furiosus
β-amylase
2xfr – bBAM
1wdp – sBAM – soybean
2dqx, 1uko, 1ukp – sBAM (mutant)
1vem, 5bca, 1cqy, 1b90 – BcBAM – Bacillus cereus
1ven - BcBAM (mutant)
1fa2 - AAM + saccharide – Sweet potato
β-amylase binary complexes
2xff – bBAM + acarbose
2xfy, 2xg9, 2xgb, 2xgi – bBAM + inhibitor
1wdq, 1wdr, 1wds, 1v3h, 1v3i, 1q6d, 1q6e, 1q6f, 1q6g - sBAM (mutant) + saccharide
1q6c, 1bfn, 1bya, 1byb, 1byc, 1byd, 1btc - sBAM + saccharide
1j0y, 1j0z, 1j10, 1j11, 1j12, 1j18, 1b9z - BcBAM + saccharide
1veo, 1vep, 1itc - BcBAM (mutant) + saccharide
1b1y - AAM (mutant) + saccharide – Hordeum vulgare
γ-amylase
1lf6 – TtGAM – Thermoanaerobacterium thermosaccharolyticum
1lf9 - TtGAM + acarbose
β/α-amylase
2laa, 2lab – PpBAAM – Paenibacillus polymyxa - NMR
[[3voc – PpBAAM
Maltohexaose-producing amylase
1wp6 - BacMAM
1wpc – BacMAM + saccharide
Maltogenic amylase
1gvi, 1sma – MAM – Thermus sp.
Taka amylase
2taa – AoTAM
7taa – AoTAM + acarbose
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press
- ↑ 2.0 2.1 Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998 Mar;7(3):564-72. PMID:9541387
- ↑ 3.0 3.1 3.2 Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability. J Biochem. 2001 Mar;129(3):461-8. PMID:11226887
- ↑ 4.0 4.1 4.2 Aghajari N, Feller G, Gerday C, Haser R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002 Jun;11(6):1435-41. PMID:12021442
- ↑ Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 6.0 6.1 Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 7.0 7.1 7.2 7.3 Kuriki T, Imanaka T. The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87(5):557-65. PMID:16232518
- ↑ 8.0 8.1 PPMID: 17713601
- ↑ Franco OL, Rigden DJ, Melo FR, Grossi-De-Sa MF. Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases. Eur J Biochem. 2002 Jan;269(2):397-412. PMID:11856298
- ↑ Yang RW, Shao ZX, Chen YY, Yin Z, Wang WJ. Lipase and pancreatic amylase activities in diagnosis of acute pancreatitis in patients with hyperamylasemia. Hepatobiliary Pancreat Dis Int. 2005 Nov;4(4):600-3. PMID:16286272
Proteopedia Page Contributors and Editors (what is this?)
Shane Riley, Michal Harel, Joel L. Sussman, Randi Woodbeck, Jaime Prilusky, Alexander Berchansky, Ann Taylor, Andrea Gorrell, David Canner