We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 828
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
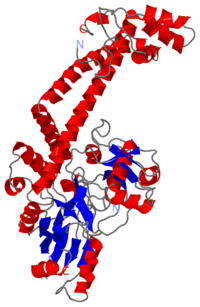
The three connecting helices (a14,a18 anda19) adopt very different conformations, leading to large quaternary movements involving a single hinge-point within the helices and rigid body movements of | The three connecting helices (a14,a18 anda19) adopt very different conformations, leading to large quaternary movements involving a single hinge-point within the helices and rigid body movements of | ||
the head fragments. | the head fragments. | ||
| + | |||
[[Image:gyrA59.jpg]] | [[Image:gyrA59.jpg]] | ||
| + | |||
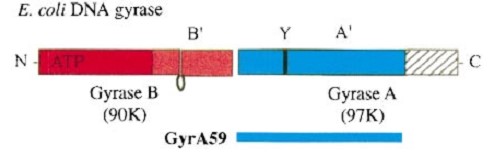
The tail is structurally conserved although large surface loops emanating from different points give it a different outward appearance It forms a heart-shaped homodimer with two protein interfaces, the DNA- and C-gates. GyrA59 is the minimal fragment of the A-subunit which, when complexed with the B-subunit, has DNA-cleavage activity. | The tail is structurally conserved although large surface loops emanating from different points give it a different outward appearance It forms a heart-shaped homodimer with two protein interfaces, the DNA- and C-gates. GyrA59 is the minimal fragment of the A-subunit which, when complexed with the B-subunit, has DNA-cleavage activity. | ||
The remaining 30–35 kDa comprising the C-terminal domain (CTD) of GyrA shows a domain forming a b-pinwheel with a positively charged amino-acid perimeter. This carboxy-terminal domain of GyrA (cyan) is required for the introduction of DNA supercoils). | The remaining 30–35 kDa comprising the C-terminal domain (CTD) of GyrA shows a domain forming a b-pinwheel with a positively charged amino-acid perimeter. This carboxy-terminal domain of GyrA (cyan) is required for the introduction of DNA supercoils). | ||
Revision as of 16:12, 24 December 2013
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||