We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 828
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
---- | ---- | ||
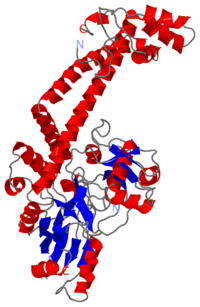
| - | <Structure load=' | + | <Structure load='1ab4_mm1.pdb' size='400' frame='true' align='right' caption='Monomer of GyrA59 subunit at 2.8Å (1ab4.pdb)' scene='56/568026/Coloured/1'> |
[[Image:1ab4.png|left|200px]] | [[Image:1ab4.png|left|200px]] | ||
| Line 34: | Line 34: | ||
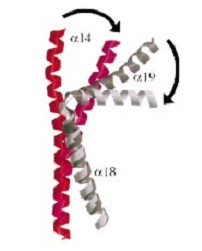
<scene name='56/568026/Helix14/1'>alpha helix 14</scene> and <scene name='56/568026/Helix18/1'>alpha helix 18</scene>) emanate from this core and connect, together with the C-terminal helix ( | <scene name='56/568026/Helix14/1'>alpha helix 14</scene> and <scene name='56/568026/Helix18/1'>alpha helix 18</scene>) emanate from this core and connect, together with the C-terminal helix ( | ||
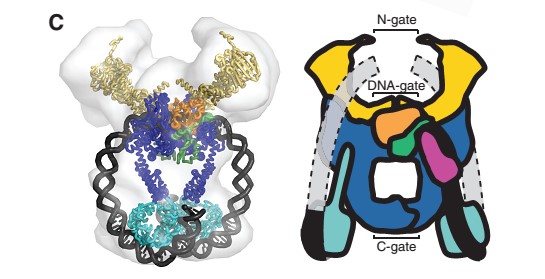
<scene name='56/568026/Helix19/1'>alpha helix 19</scene>), the head and tail fragments. This domain has a small globular domain at it's end and is involved in the dimerization creating the <scene name='56/568026/Coiledcoil/1'>C-gate</scene> . | <scene name='56/568026/Helix19/1'>alpha helix 19</scene>), the head and tail fragments. This domain has a small globular domain at it's end and is involved in the dimerization creating the <scene name='56/568026/Coiledcoil/1'>C-gate</scene> . | ||
| - | |||
| - | |||
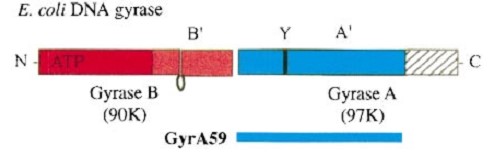
| - | [[Image:gyrA59.jpg]] | ||
| Line 79: | Line 76: | ||
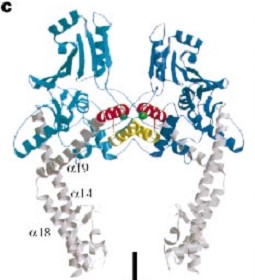
The gyrase structure reveals a '''new cluster of conserved residues''', juxtaposing Tyr 122 and Arg 121 from one monomer and His 80, Arg 32 and Lys 42 from the other monomer. '''This cluster may form the active site of the breakage–reunion reaction''', with the other conserved positive charges (Arg 46 and Arg 47) anchoring the non-covalently bound 3' end of the cleaved DNA. | The gyrase structure reveals a '''new cluster of conserved residues''', juxtaposing Tyr 122 and Arg 121 from one monomer and His 80, Arg 32 and Lys 42 from the other monomer. '''This cluster may form the active site of the breakage–reunion reaction''', with the other conserved positive charges (Arg 46 and Arg 47) anchoring the non-covalently bound 3' end of the cleaved DNA. | ||
| - | <Structure load='1ab4_mm1.pdb' size='300' frame='true' align='right' caption='GyrA59, Breakage/Reunion subunit (dimer)' scene='56/568026/Biounit/1' /> | ||
Revision as of 14:47, 29 December 2013
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||